Bacteria Raman spectrum identification and classification method based on SIMCA-SVDD
A technology of Raman spectroscopy, recognition and classification, applied in the directions of Raman scattering, character and pattern recognition, material excitation analysis, etc. species identification etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0066] The present invention is described in detail in conjunction with the contents and embodiments of the accompanying drawings.
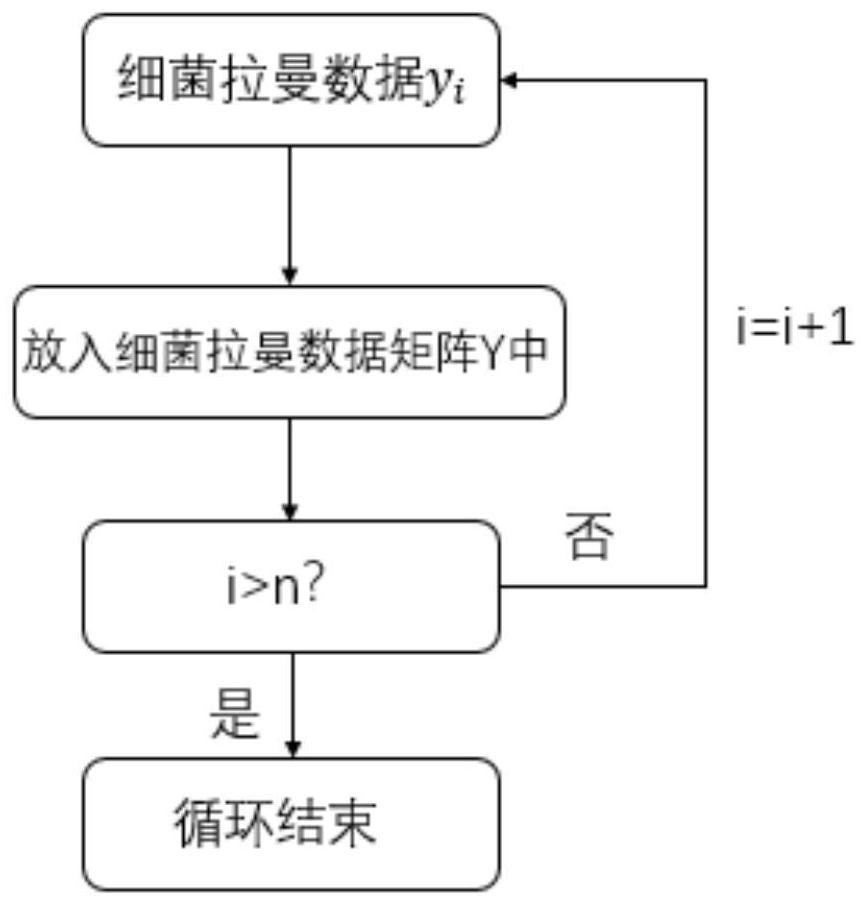
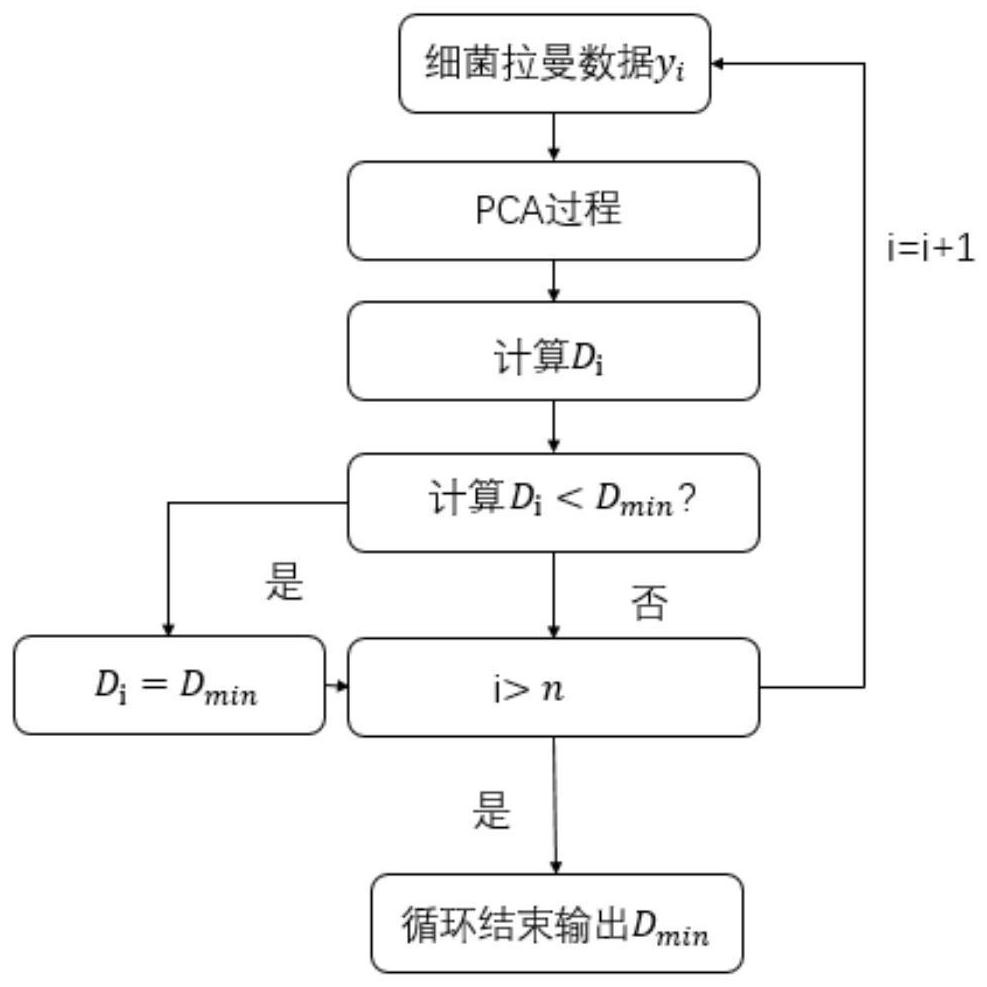
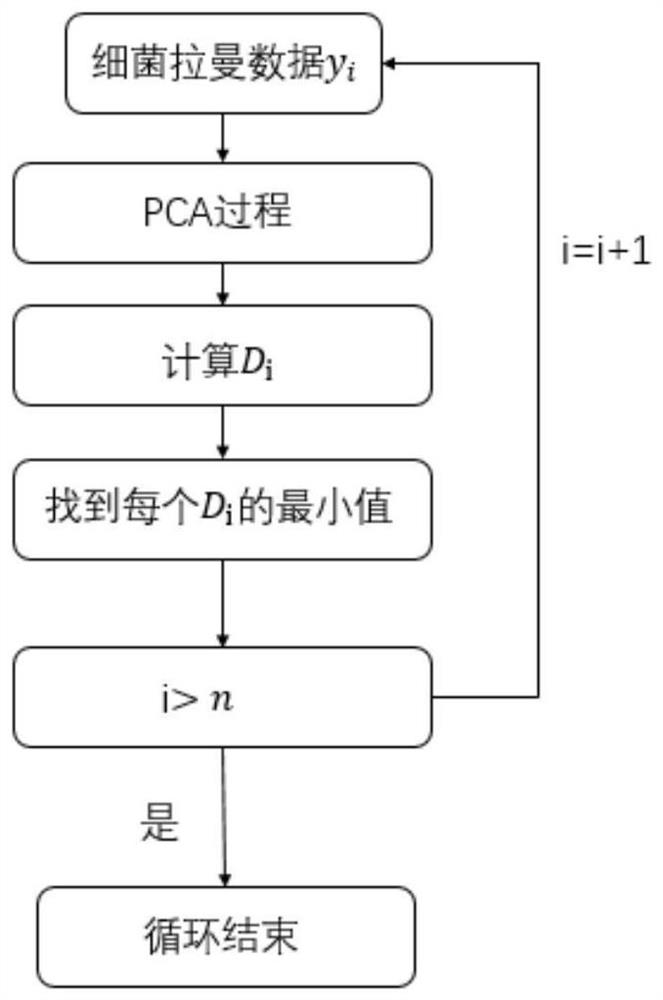
[0067] like Figure 1-5 As shown, the present invention provides a SIMCA-SVDD-based bacterial Raman spectrum identification and classification method. In order to make the purpose, technical solutions and effects of the present invention clearer and clearer, the present invention will be further described in detail below. It should be understood that the specific embodiments described in the present invention are only used to explain the present invention, and are not intended to limit the present invention.
[0068] from Figure 4-Figure 5 It can be seen that the original SIMCA and SIMCA-SVDD methods have achieved 100% accuracy in the identification and classification of Escherichia coli, Shigella flexneri, and Clostridium difficile. However, when identifying and classifying Clostridium botulinum, the accuracy rate of SIMCA was 86.7%, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com