Methods for preparing nucleic acid molecules for sequencing
A sequencing and nucleotide technology, applied in the field of rolling circle amplification, can solve problems such as high error rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
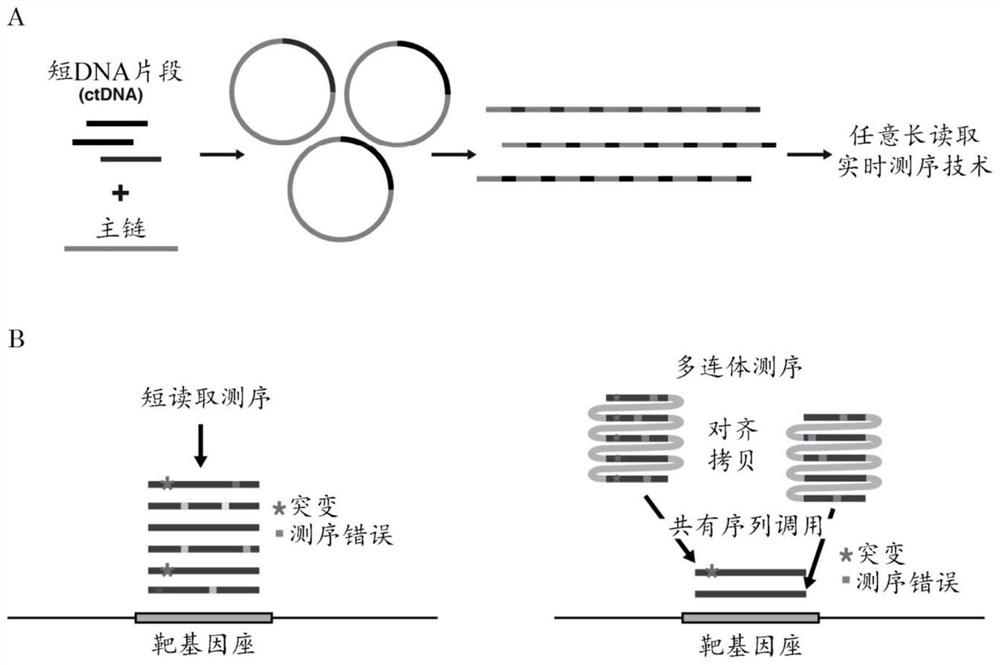
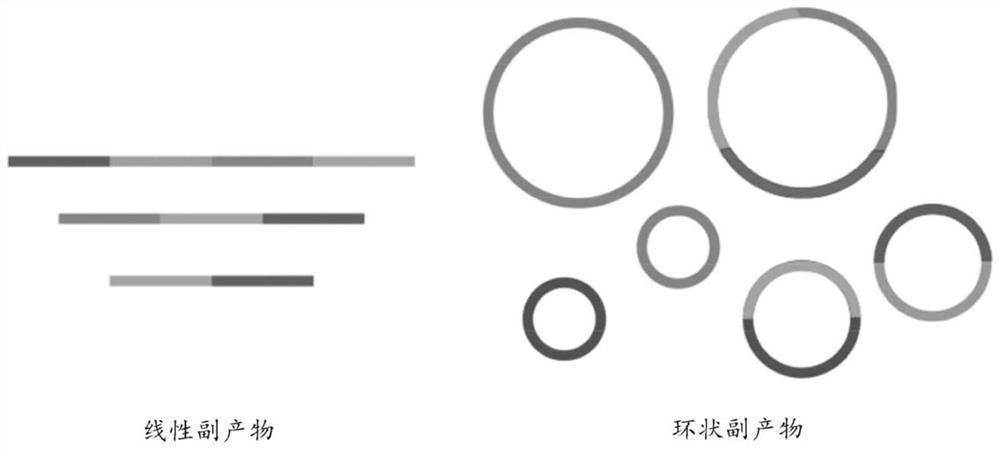
[0046] The means and methods described herein allow multiple determinations of the sequence of the same target DNA molecule. This can be used as a means of correcting errors. This differs from classical second-generation sequencing approaches that correct for errors by sequencing multiple independent molecules covering the same genomic locus. In this case, each read typically represents one sequencing event for one molecule. With the methods of the invention, a single (target) molecule is replicated repeatedly so that a single read represents multiple sequencing events for the same molecule.
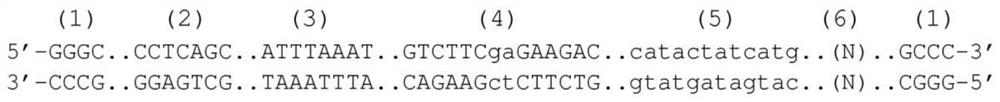
[0047] The target nucleic acid is usually double-stranded DNA. Single-stranded DNA or RNA for which a defined sequence is desired can be readily converted to double-stranded DNA by methods known in the art. Such methods include, but are not limited to, cDNA synthesis, reverse transcriptase (RT) polymerase chain reaction (PCR), PCR, random primer extension, and the like. The target DN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com