Method for detecting interaction between biofilm proteins and complete set of reagents used by method
A protein and biofilm technology, applied in biological testing, using carriers to introduce foreign genetic material, material inspection products, etc., can solve problems such as excessive false positive results, affecting the effect of immunoprecipitation, and affecting the results of activity experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
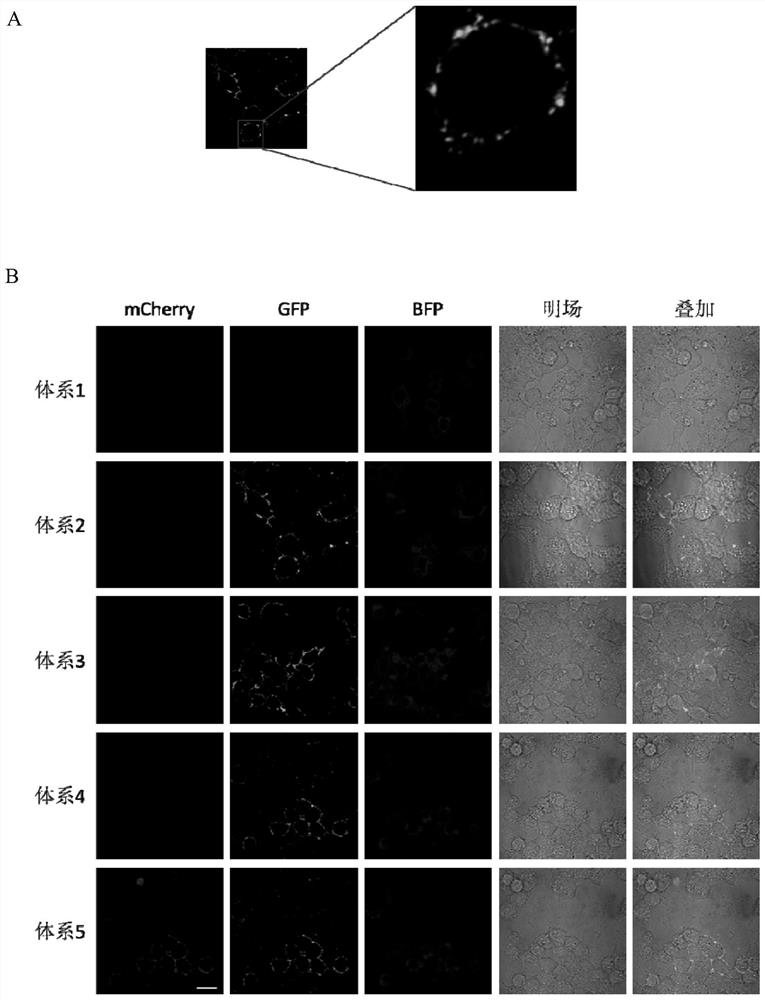
[0146] Example 1, identification of cell membrane protein interaction promoting factors
[0147] 1. Preparation of recombinant vector
[0148] 1. A recombinant vector expressing the fusion protein of mCherry and FRB
[0149] The DNA fragment (comprising the recognition sequence of NcoI and XhoI) between the NcoI and XhoI recognition sequences of the pRSFDuet-1 vector (Novagen product of Merck) is replaced with the DNA molecule shown in the 11th-831th position of sequence 2 in the sequence listing, The recombinant vector pRSFDuet-1-mCherry is obtained, and the pRSFDuet-1-mCherry can express the protein shown in Sequence 1 (mCherry is fused with His-tag, denoted as His-mCherry).
[0150] Among them, the 13th-825th position of sequence 2 encodes His-mCherry shown in sequence 1, the 815-820th and 826-831st positions of sequence 2 are the recognition sequences of NcoI and XhoI respectively, and the 3rd-8th position of sequence 1 The position is the amino acid sequence of His-tag,...
Embodiment 2
[0201] Example 2, verification of cell membrane protein interaction inhibitors
[0202] 1. Preparation of recombinant vector
[0203] 1. A recombinant vector expressing the fusion protein of mCherry and KKETPV
[0204] The DNA fragment between the NcoI and XhoI recognition sequences of the pRSFDuet-1-mCherry vector of Example 1 (comprising the recognition sequences of NcoI and XhoI) was replaced with the DNA molecule shown in sequence 8 to obtain the recombinant vector pRSFDuet-1-mCherry-KKETPV , pRSFDuet-1-mCherry-KKETPV expresses the fusion protein of His-mCherry and KKETPV shown in Sequence 7 (the fusion protein is designated as mCherry-KKETPV, and contains a His tag).
[0205] 2. A recombinant vector expressing the fusion protein of GFP-LCD and FRB
[0206]The DNA fragment between the NcoI and XhoI recognition sequences of the pRSFDuet-1-GFP-LCD vector of Example 1 (comprising the recognition sequences of NcoI and XhoI) was replaced with the DNA molecule shown in sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com