Method for identifying driver gene from differential network
A driver gene and network technology, applied in the computer field, can solve problems such as insufficient accuracy of driver gene identification, failure to consider gene regulation network uncertainty, and inability to comprehensively measure network topology differences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
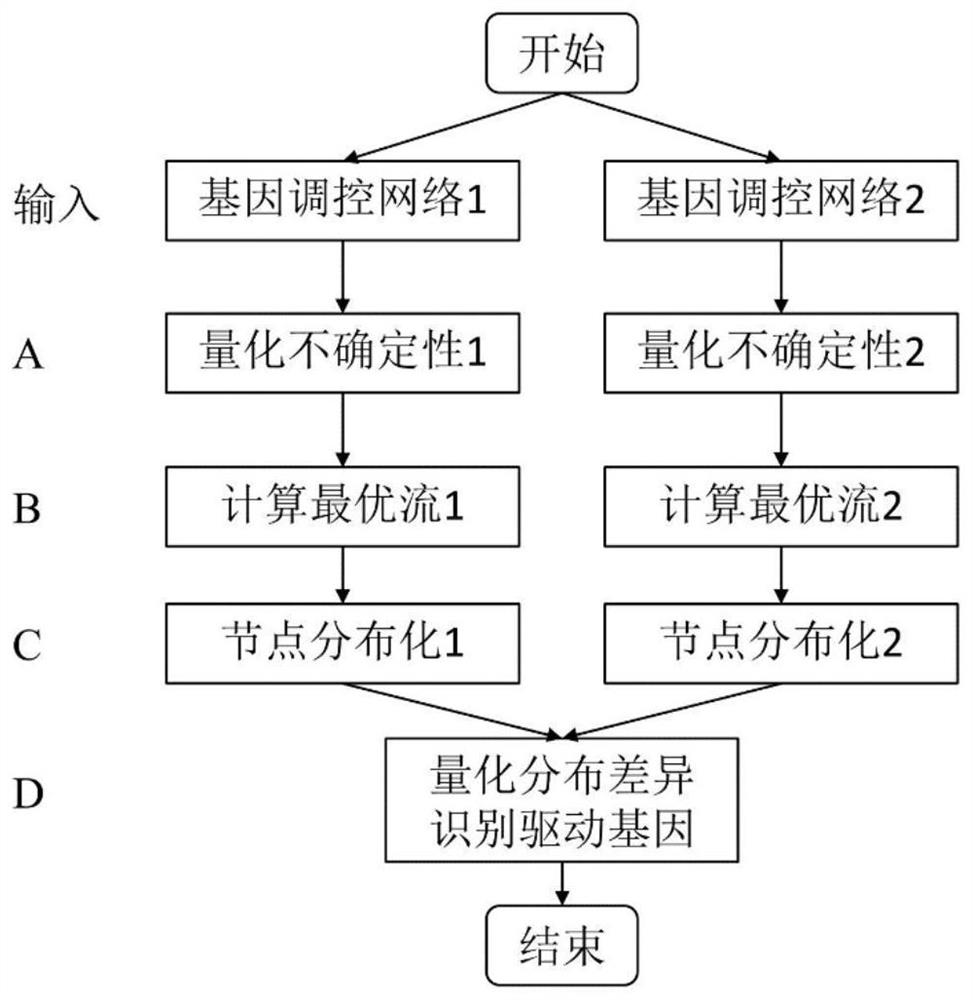
[0043] Preferred embodiments of the present invention will be further described in detail below in conjunction with the accompanying drawings.
[0044] In this embodiment, the DNF algorithm is implemented using C++ and R language, and the hardware environment of all experiments is a notebook computer with an Intel Core I5 main frequency of 1.8GHz, 8GB of memory and a 64-bit Windows 10 operating system.
[0045] The first is the construction of the difference network model. From the computer point of view, given a gene regulatory network with n nodes, the network A_(i,j),i,j=1,...,n can be used to describe the relationship between any pair of genes. connection between. In a directed gene regulatory network, for a given gene i and gene j, their relationship in the network is described as A_(i,j)=A_(i→j)=c, which means a directed connection from gene i Extending to gene j, its action strength coefficient is c; in the undirected gene regulation network, for a given gene i and g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com