Primer set for detecting human SERPINF1 gene mutation and kit thereof
A kit and genome technology, applied in the field of primer sets and kits for detecting human SERPINF1 gene mutations, can solve problems such as missing test results, failure to detect large fragment deletions, copy number variations, and no clear candidate genes, etc., to achieve Avoid omissions, save costs and manpower, and ensure accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
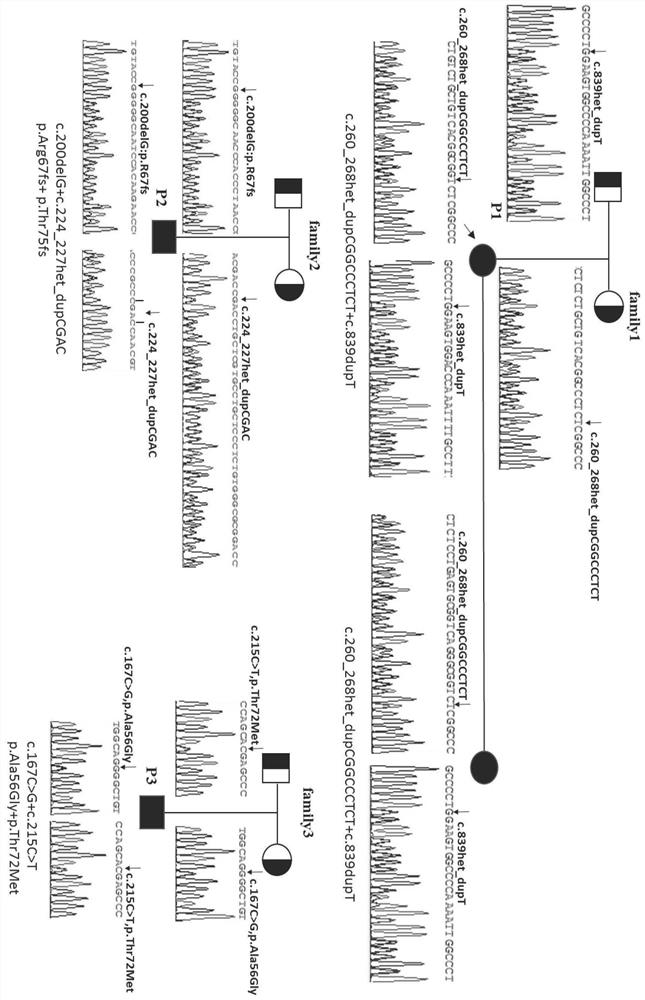
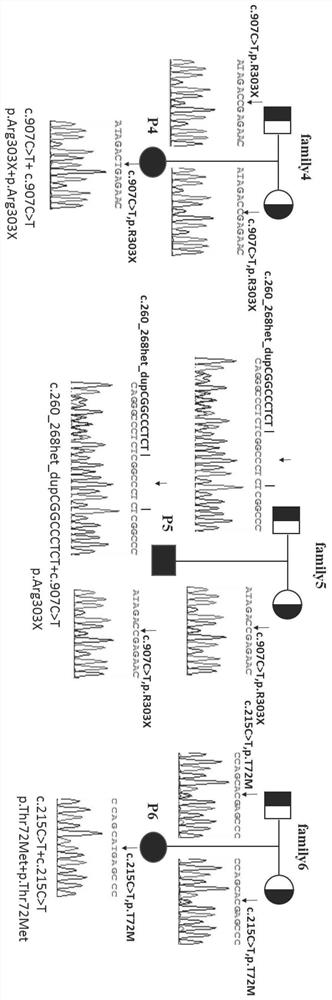
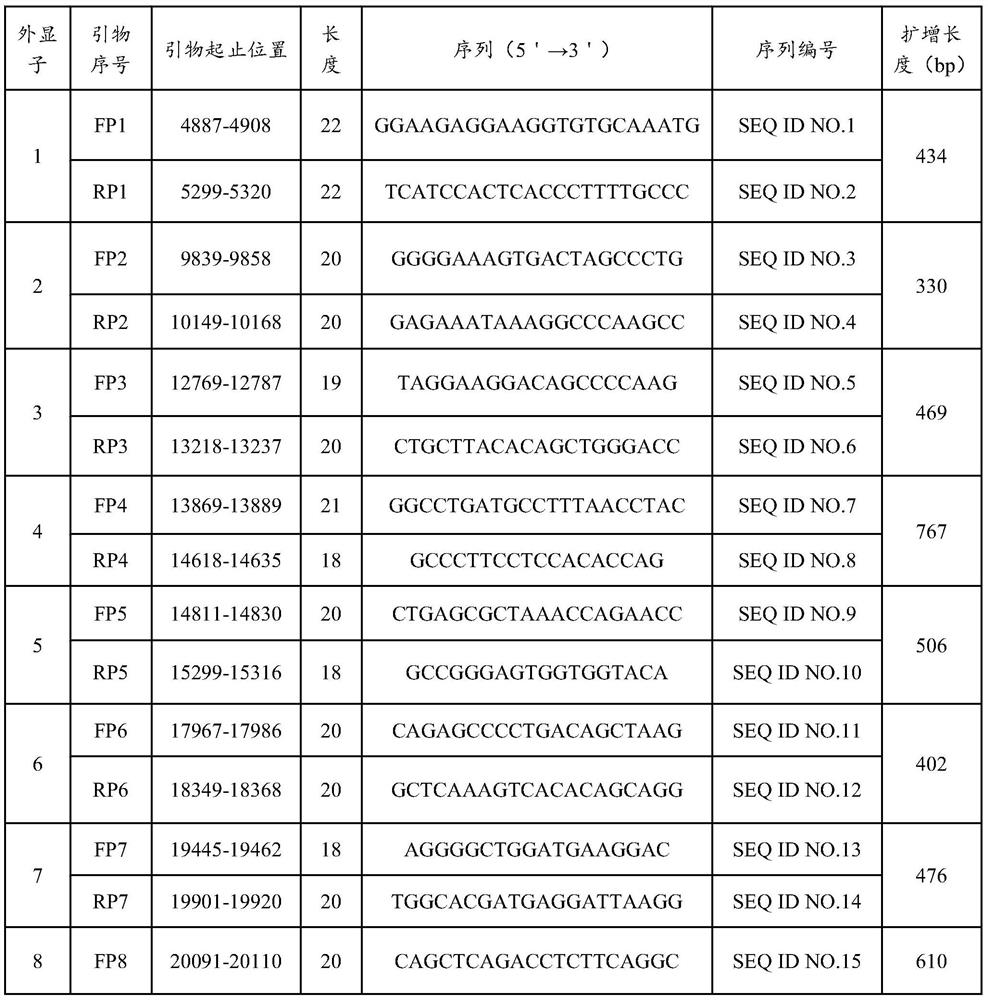
[0063] The DNA sequence of the SERPINF1 gene (NG_028180.1) was analyzed according to the NCBI GeneBank database, and the amplification primers for exons 1 to 8 were designed. The amplified region includes all exons of SERPINF1 and the intron sequence at the junction of exons and introns not less than 50 bp. The nucleotide sequences of primers used in PCR amplification are shown in Table 1.
[0064] Table 1 SERPINF1 gene exon primer amplification and sequencing sequence
[0065]
[0066]
Embodiment 2
[0068] A kit for detecting SERPINF1 gene mutation, comprising:
[0069] The primer set for PCR amplification from the 1st to the 8th exon designed in Example 1 for detecting the mutation of the SERPINF1 gene;
[0070] PCR amplification reagents include: 10mM dNTP Mix, containing Mg 2+ 2×Phanta Max Buffer, Phanta MaxSuper-Fidelity DNA Polymerase and de-RNase and DNase enzyme water (DEPC water); the working concentration of each component in the PCR amplification system is: 10000μM dNTP Mix / 2mM 2×Phanta Max Buffer / 1U / μl Phanta MaxSuper-Fidelity DNA Polymerase, the upstream and downstream primers are 5μM and the template concentration is 50-400ng / μl.
[0071] PCR product purification reagents include: 1U / μl SAP enzyme, 10U / μl ExoI enzyme, RNase and DNase-free water; the working concentration of each component is: 0.05U / μl SAP enzyme, 0.5U / μl ExoI enzyme.
[0072] DNA sequencing reagents include: the above sequencing primer set for detecting SERPINF1 gene mutation, BigDye 3.1 m...
Embodiment 3
[0076] Utilize the kit for detecting the mutation of the SERPINF1 gene in the above-mentioned embodiment 2 to carry out the detection method of the mutation of the human SERPINF1 gene, the steps are as follows:
[0077] (1) Extraction of genomic DNA from peripheral blood
[0078]Collect 5ml of peripheral blood from the patient, use Omega Company DNA Extraction Kit (D3392-01) to extract whole blood genomic DNA, and use NanoDrop 2000 / 2000c spectrophotometer to measure DNA concentration and purity; see the product manual for specific operating steps and conditions;
[0079] (2) PCR amplification
[0080] The PCR reaction system is 40 μl, including:
[0081] 10mM dNTP Mix 1μl, 2×Phanta Max Buffer 20μl, Phanta Max Super-Fidelity DNA Polymerase 1μl, upstream primer 4μl, downstream primer 4μl, DNA template 50ng~400ng, make up to 40μl with DEPC water.
[0082] Use the PIC-200 PCR instrument of BIO-RAD Company to perform touchdown PCR. The PCR reaction conditions are as follows: 95°C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com