Microbial traceability method for pig manure pollution
A technology for microorganisms and pig feces, which is applied in the field of microbial traceability of pig feces pollution, can solve the problems of false positives, interference of microbial traceability detection, and false negative test results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
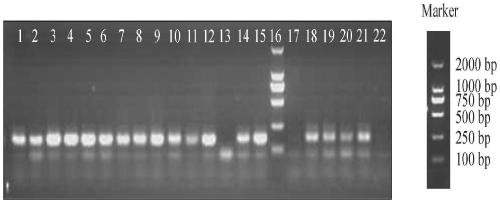
[0033] Sample DNA Extraction
[0034] Fecal samples were directly used with FastDNA TM Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) kit was used to extract DNA; after the environmental water samples were filtered through a 0.45 μm vacuum filter, the microorganisms were all trapped on the filter membrane, the filter membrane was cut into pieces, and then used FastDNA TMSpin Kit for Soil (MP Biomedicals, Solon, OH, USA) kit was used to extract DNA.
Embodiment 2
[0036] Primer screening:
[0037] 1 set of primers:
[0038] Forward primer PF163F: 5'-GCGGATTAATACCGTATGA-3', shown in SEQ ID NO:1;
[0039] Reverse primer Bac708R: 5'-CAATCGGAGTTCTTCGTG-3', shown in SEQ ID NO:2.
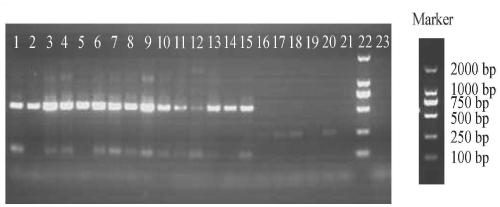
[0040] The target microorganism is porcine-specific Bacteroides, and the target fragment is a partial fragment of the 16S rRNA gene, with a size of 563bp.
[0041] 2 sets of primers:
[0042] Forward primer PS422F: 5'-CGGGTTGTAAACTGCTTTTATGAAG-3', shown in SEQ ID NO:3;
[0043] Reverse primer Bac581R: 5'-CGCTCCCTTTAAACCCAATAAA-3', shown in SEQ ID NO:4.
[0044] The target microorganism is porcine-specific Bacteroides, and the target fragment is a partial fragment of the 16S rRNA gene, with a size of 150bp.
[0045] 3 sets of primers:
[0046] Forward primer Bac41F: 5'-TACAGGCTTAACACATGCAAGTCG-3', shown in SEQ ID NO:5;
[0047] Reverse primer PS183R: 5'-CTCATACGGTATTAATCCGCCTTT-3', shown in SEQ ID NO:6.
[0048] The target microorganism is porcine-specific B...
Embodiment 3
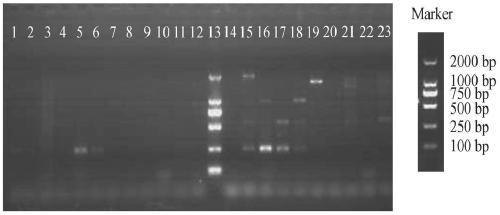
[0073] Screening of reaction conditions
[0074] One set of primers in Example 2 was used, the reaction system remained unchanged, and the annealing temperature was increased by gradient (1°C) to screen for the most suitable reaction conditions.
[0075] The results showed that the best reaction conditions for PCR were: 95°C for 3min, 30 cycles: 95°C for 30s, 62°C for 1min, 72°C for 30s, and finally 72°C for 10min.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com