Microsatellite instability prediction method and application thereof
A technology of microsatellite instability and prediction method, which is applied in biochemical equipment and methods, microbial determination/inspection, genomics, etc., and can solve the problems of uneven detection standards and unsatisfactory performance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
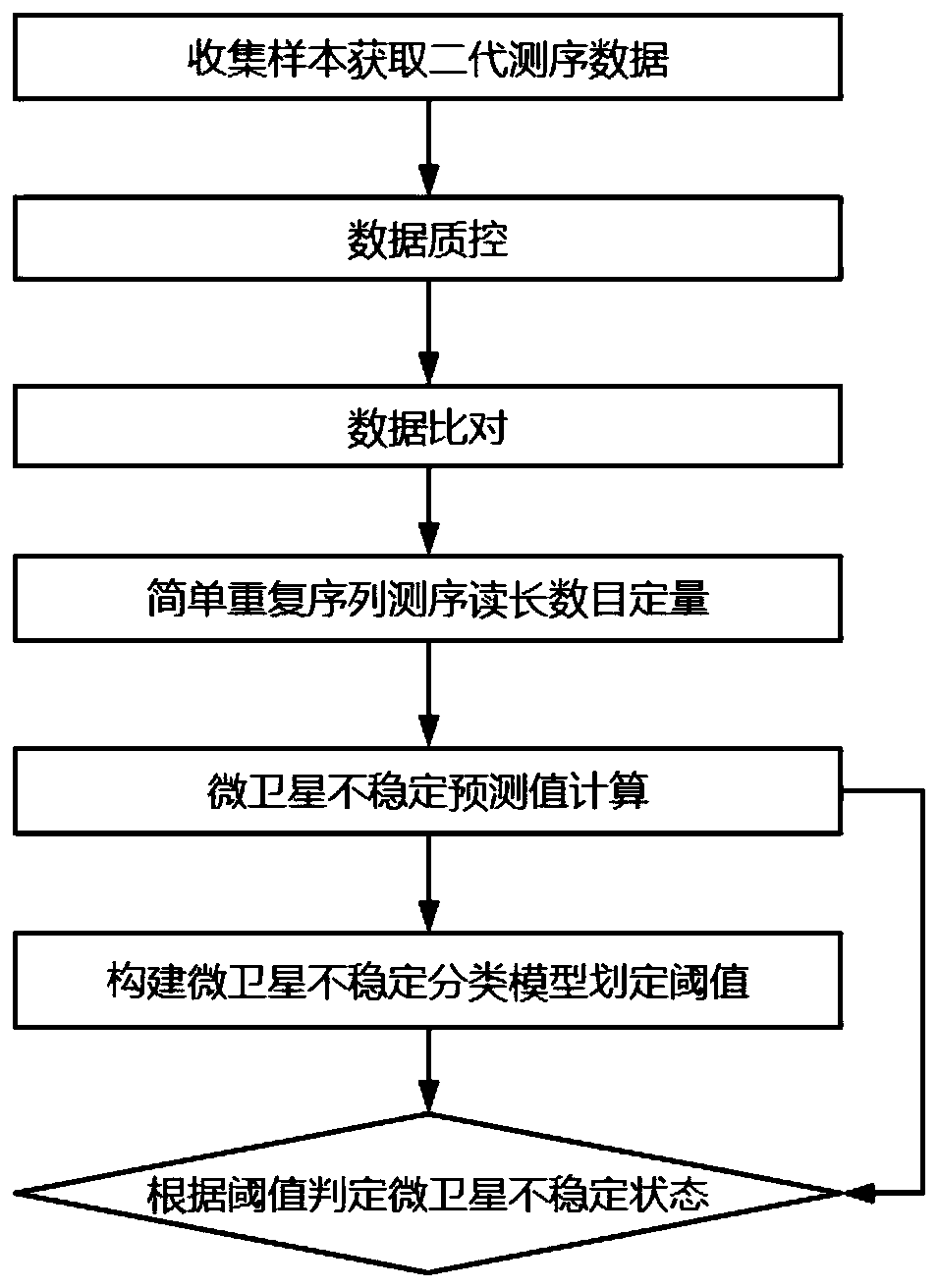
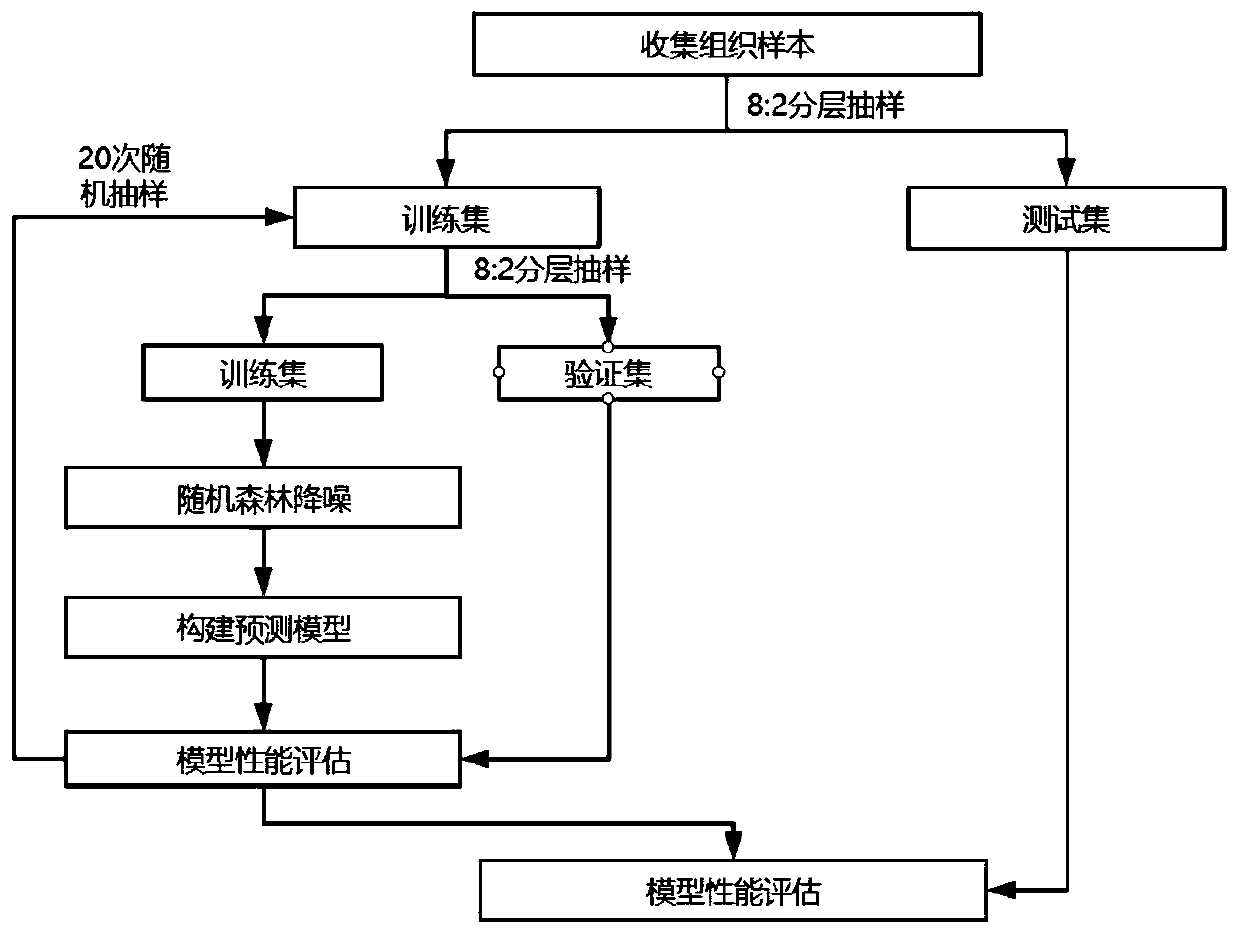
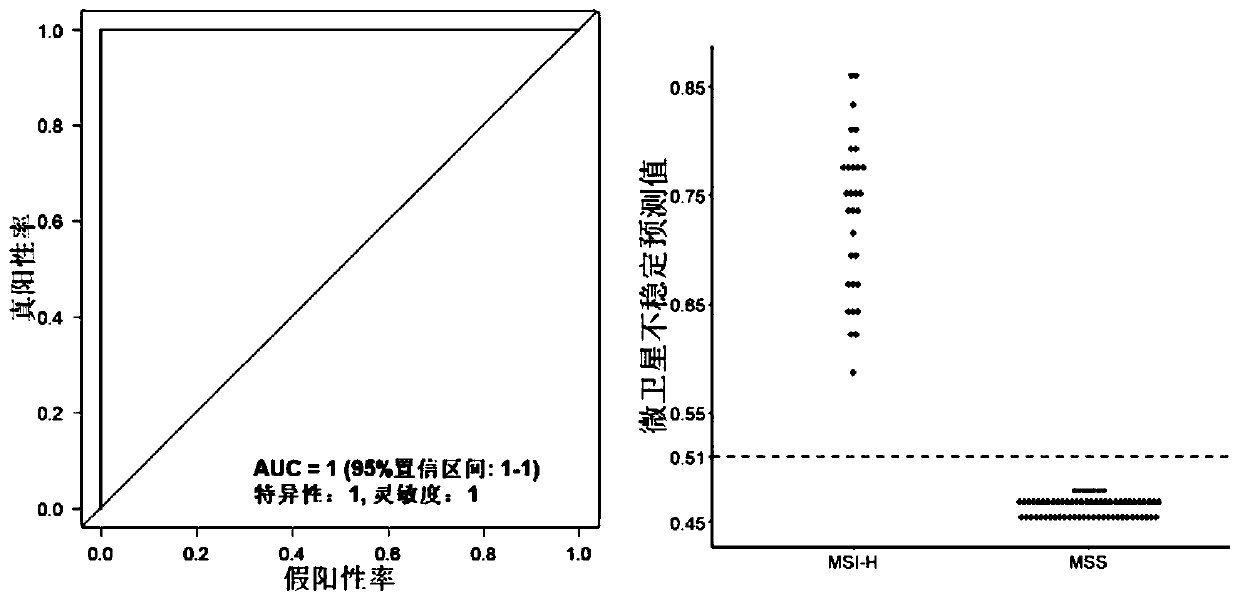
[0097] Collect clinical patient tissue samples, including tumor samples to be tested and their paired healthy control (normal) samples, for testing the MSI detection method of the present invention, and verify it with PCR MSI detection method, and compare with other existing technologies Known MSI detection tools for comparison. Wherein, the PCR MSI detection result is used as the gold standard to compare its consistency with the result of the MSI detection method of the present invention to evaluate the performance of the MSI detection method of the present invention. It should be noted that the MSI detection method proposed in the present invention does not require the use of control samples, which are used for PCR verification and other MSI detection software analysis in the Examples section.
[0098] As for the MSI loci used for testing, a large gene panel was used in this example for testing. The panel contained 654 genes related to tumors such as rectal cancer, gastric c...
Embodiment 2
[0101] Exon probe capture library construction sequencing was performed on the 4 gene combinations in Example 1, and a gene sequencer (NextSeq CN500) was used to perform 150bp Pair-End mode sequencing (Read1: 151, Read2: 151) according to the standard operating procedures of the instrument. ; Index1:8, Index2:8), and finally get the next-generation sequencing data in fastq format as raw data (rawdata).
[0102] The quality control software fastp was used to control the quality of the obtained next-generation sequencing data, and filter the sequencing joints, low-quality bases and sequencing error fragments, etc., and obtain high-quality data (clean data) after filtering.
[0103] Use the comparison software bwa to compare the clean data with the reference genome hg19 to obtain the corresponding specific position information on the genome of each DNA fragment; then use the gencore software to compare and de-duplicate and base-correct the data.
[0104] According to the repeat (...
Embodiment 3
[0106] According to the repeatcount result obtained in Example 2, all repeat types obtained at each site represent the allele type of the position, and for each loci, the difference between the main allele and the secondary allele abundance values is calculated as the difference between the loci microsatellites Stable level prediction value, the average value of all loci in the sample (1-unstable level prediction value) is used as the microsatellite instability prediction value (MSIscore) of the sample, and the calculation formula of the non-parametric algorithm is as follows:
[0107]
[0108] Among them, X i1 Indicates the main peak, X i2 Indicates the secondary peak, i is the order of loci, and n is the number of loci.
[0109] Specifically, count the repeatcount of each detection position with a repeat length between 0-50bp, and normalize the repeatcount so that it falls within the range of 0-1. The normalization method is the reads of each repeat type at this site ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com