Novel DNA (deoxyribonucleic acid) nucleic acid cleaving enzyme and application thereof
A polynucleotide and nucleic acid construct technology, applied in the field of new DNA nuclease cutting enzymes, can solve the problems of inability to recognize target genes and low editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0264] Example 1 Editing experiments in tomato of expression vectors containing different Cas9 variants
[0265] 1. Experimental materials
[0266] Tomato (Solanum lycopersicum cultivar Ailsa Craig), vector pCAMBIA1300, restriction enzymes EcoRI and HindIII, T4 ligase
[0267] 2. Carrier construction
[0268] Use the PCR method to mutate the corresponding mutation site, and connect it to the corresponding vector using double enzyme digestion and T4 ligase. For the structure of the editing tool, see figure 1 a.
[0269] 3. Experimental method
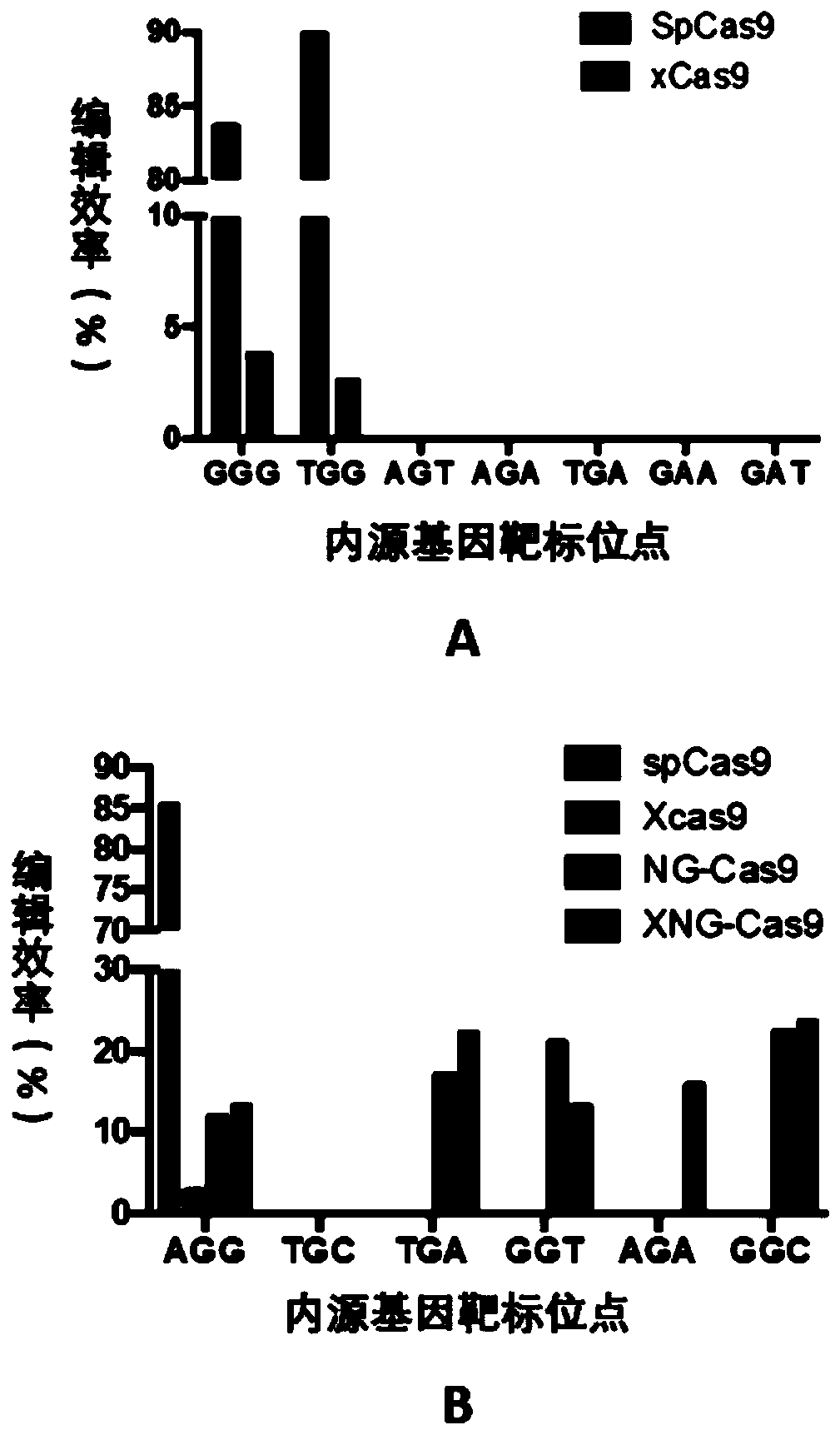
[0270] Two target genes SlBRI1 (regulating plant type) and SlRIN (regulating fruit ripening) in tomato were selected, and 11 sgRNAs were designed for these two genes, including NGG PAMs (AGG, TGG and GGG) and NG PAMs (TGC, TGA , GGT, AGA, GGC, AGT) (see Table 1), for each sgRNA, 40T1 transgenic tomato plants were generated respectively.
[0271] Table 1 Primers used to generate sgRNA plasmids and sequence in tomato
[0272]
[0...
Embodiment 2
[0288] Example 2 Editing experiment of expression vectors containing different Cas9 variants and ABE in tomato
[0289] 1. Plant material
[0290] Wild-type tomato (variety, Ailsa Craig), all transfected tomatoes were grown in a standard greenhouse (12 hours light at 26°C / 12 hours light at 20°C)
[0291] 2. Carrier Construction
[0292] PCR was used to complete the cloning of the coding region of ABE 7.10 (TadA-TadA7.10), and PCR was used to complete gene fusion and gene mutation. The promoters used were RPS5A, Pro35S, and AtU6. The primers are shown in Table 2, and the structure of the editing tool is shown in figure 1 b.
[0293] 3. Tomato transformation
[0294] The binary plasmid expressing sgRNA and ABE was transfected into Agrobacterium agrobacterium EHA105, and the tomato cotyledons were infected with the Agrobacterium.
[0295] 4. Mutation detection
[0296] DNA from tomato leaves was extracted by the CTAB method, and 14 gene regions containing sgRNA target sites...
Embodiment 3
[0300] Example 3 Editing experiment of expression vectors containing different Cas9 variants in Arabidopsis
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com