Cell apoptosis gene segment for reducing egg laying amount of small brown rice planthopper and application thereof
A gene fragment, the technology of the small planthopper, applied in the field of agricultural science, achieves the effect of facilitating large-scale popularization and application, reducing the amount of eggs laid, and the amount of eggs laid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
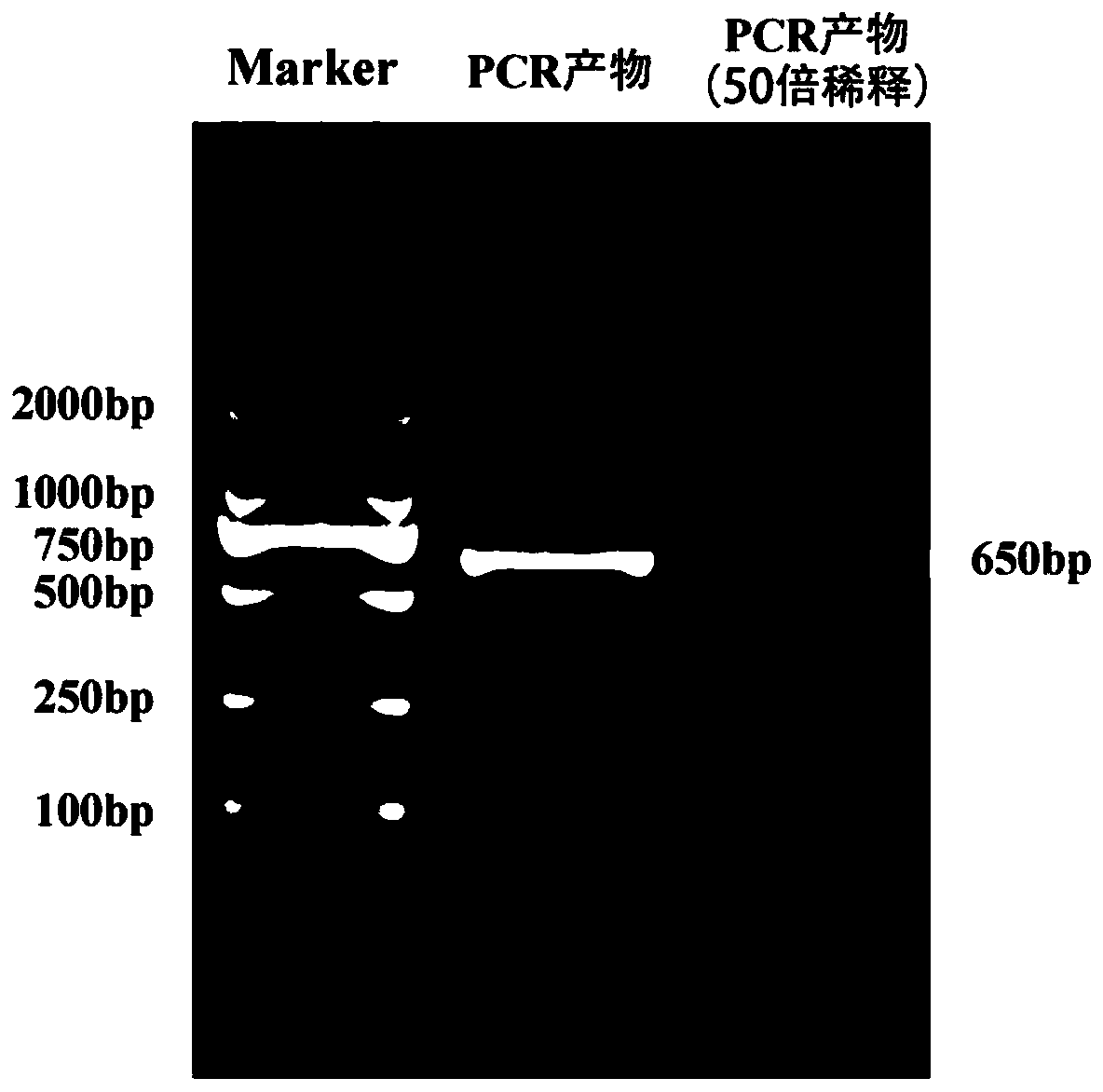
[0032] Example 1, the method for cloning the Lsbcl-2 fragment of the L. striatellus cell apoptosis gene.
[0033] Specific steps are as follows:
[0034] Step 1, take 40-50 ovaries of female adult SBPH, and extract total RNA by TRizol method;
[0035] Step 2, using total RNA as a template, reverse transcription to synthesize the first strand of cDNA;
[0036] Step 3: Obtain gene fragment sequences from the SBPH genome, and perform homology comparisons in the NCBI database (http: / / www.ncbi.nlm.nih.gov / ) to predict cell apoptosis in SBPH For the gene Lsbcl-2, the upstream primer P1 (SEQ ID NO.3) and the downstream primer P2 ( SEQ ID NO.4), carry out PCR amplification;
[0037] Upstream primer P1 (SEQ ID NO.3): 5'-CGTCTGCTGAAGTGGTGGAT-3'
[0038] Downstream primer P2 (SEQ ID NO.4): 5'-TCAGGAAACACCTCTCGCAC-3'
[0039] The PCR amplification system is as follows: 4 μL 5× reaction buffer buffer, 2 μL dNTP, 1 μL cDNA template in step (1), 2 μL upstream primer P1 whose sequence is...
Embodiment 2
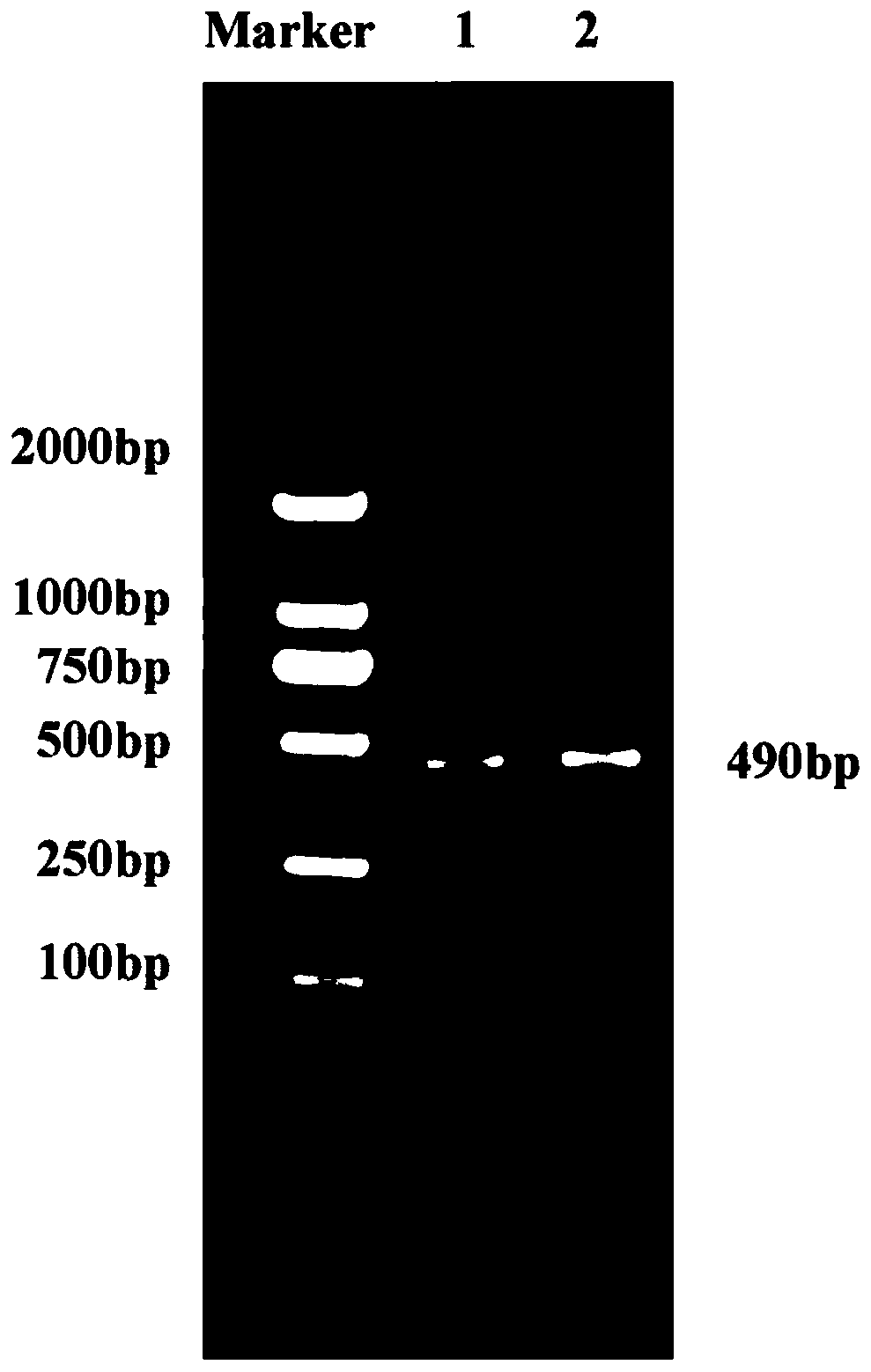
[0041] Example 2, the dsRNA synthesis and recovery of the apoptosis gene Lsbcl-2 of SBPH.
[0042] Specific steps are as follows:
[0043] Step 1. Based on the verified gene sequence, use Primer-Blast in the NCBI database (https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ) to design the upstream primer P3 (SEQ ID NO.6) and downstream primer P4 (SEQ ID NO.7)
[0044] Upstream primer P3 (SEQ ID NO.6):
[0045] 5′-taatacgactcactatagggGTGAGCGATTTGCCTTTGGG-3′
[0046] Downstream primer P4 (SEQ ID NO.7):
[0047] 5′-taatacgactcactatagggGTAGACCGCCATCCGATTGT-3′
[0048] Control GFP gene design upstream primer P5 (SEQ ID NO.8) and downstream primer P6 (SEQ ID NO.9)
[0049] Upstream primer P5 (SEQ ID NO.8):
[0050] 5′-taatacgactcactatagggAGAATGAGTAAAGGAGAAGAACTTTTC-3′
[0051] Downstream primer P6 (SEQ ID NO.9):
[0052] 5′-taatacgactcactatagggAGATTTGTATAGTTCATCCATGCCATGT-3′
[0053] The PCR amplification system is as follows: 10 μL 2× reaction buffer buffer, 2 μL upstream prime...
Embodiment 3
[0070] Example 3, experiment of feeding dsRNA on SBPH.
[0071] Specific steps are as follows:
[0072] Step 1, uniformly mixing the SBPH feed with the dsRNA (concentration of 4000 ng / μL) of the apoptosis gene;
[0073] Step 2, prepare a plurality of double-pass glass tubes, seal one end of the double-pass glass tubes with gauze and rubber bands, and use an electric insect sucker to suck 2-3 instar SBPH nymphs into the double-pass glass tubes;
[0074] Step 3: Seal the other end of the double-way glass tube with Parafilm, and use a pipette to draw 100 μL of dsRNA mixture of SBPH feed and apoptosis gene (treatment group) or 100 μL of SBPH feed and control GFP gene The dsGFP mixture (control group) was dropped in the center of the parafilm, and another parafilm was used to seal the mixture of SBPH feed and apoptosis dsRNA between the two layers of parafilm;
[0075] Step 4, place the double-pass glass tube filled with the mixture of SBPH feed and apoptosis dsRNA at a temperatu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com