Methods and compositions for improving plant traits
一种植物、植物根的技术,应用在用于改良植物性状及组合物领域,能够解决途径不清楚等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0351] Example 1: Isolation of microorganisms from plant tissue
[0352] Topsoil was obtained from various agricultural regions in central California. Twenty plots of soil with different textural characteristics were collected, including heavy clay, peat clay loam, silty clay and sandy loam. Seeds of various field corn, sweet corn, conventional corn and tomato were planted in each soil as shown in Table 1.
[0353]
[0354] Table 1: Types and varieties of crops grown in soils with different characteristics
[0355] Plants were pulled out after 2-4 weeks of growth, and excess soil on the root surfaces was removed with deionized water. After removing the soil, surface-sterilize the plant with bleach and rinse vigorously in sterile water. Cleaned 1 cm root segments were excised from plants and placed in phosphate-buffered saline solution containing 3 mm steel beads. A slurry was generated by vigorously shaking the solution with a Qiagen TissueLyser II.
[0356] Root and...
Embodiment 2
[0358] Example 2: Characterization of isolated microorganisms
[0359] Sequencing, analysis and phylogenetic characterization
[0360] 16S rDNA sequencing with the 515f-806r primer set was used to generate preliminary phylogenetic characterization of isolated and candidate microorganisms (see eg Vernon et al.; BMC Microbiol. 2002 Dec 23; 2:39). As shown in Table 2, microorganisms include different genera including: Enterobacter, Burkholderia, Klebsiella, Bradyrhizobium, Lahnella, Xanthomonas genera, Raoultella, Pantoea, Pseudomonas, Brevundomonas, Agrobacterium, and Paenibacillus.
[0361]
[0362]
[0363] Table 2: Diversity of microorganisms isolated from tomato plants determined by deep 16S rDNA sequencing.
[0364] Subsequently, the genomes of 39 candidate microorganisms were sequenced using the Illumina Miseq platform. Genomic DNA from pure cultures was extracted using the QIAmp DNA Mini Kit (QIAGEN), and total DNA libraries for sequencing were prepared by a th...
Embodiment 3
[0413] Example 3: Mutagenesis of Candidate Microorganisms
[0414] λ-Red-mediated knockout
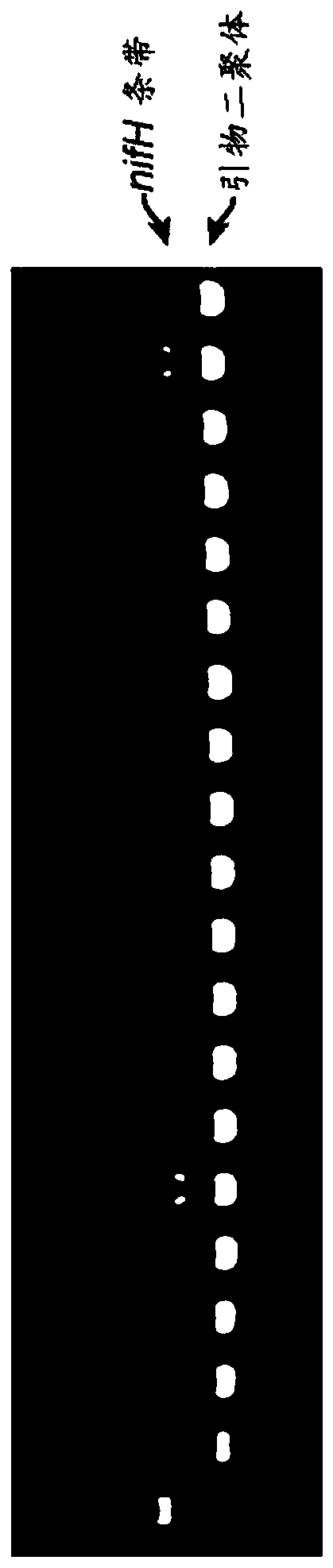
[0415] Several mutants of candidate microorganisms were generated using plasmid pKD46 or derivatives containing a kanamycin resistance marker (Datsenko et al., 2000; PNAS 97(12):6640-6645). Knockout cassettes were designed with 250 bp of homology flanking the target gene and generated by overlap extension PCR. Candidate microorganisms were transformed with pKD46, grown in the presence of arabinose to induce expression of the λ-Red machinery, ready for electroporation, and transformed with the knockout cassette to generate candidate mutagenized strains. As shown in Table 4, four candidate microorganisms and one laboratory strain Klebsiella oxytoca M5A1 were used to generate thirteen candidate mutants of the nitrogen fixation regulatory genes nifL, glnB and amtB.
[0416]
[0417] Table 4: List of individual knockout mutants generated by λ-Red mutagenesis
[0418] Oligonucleotide...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com