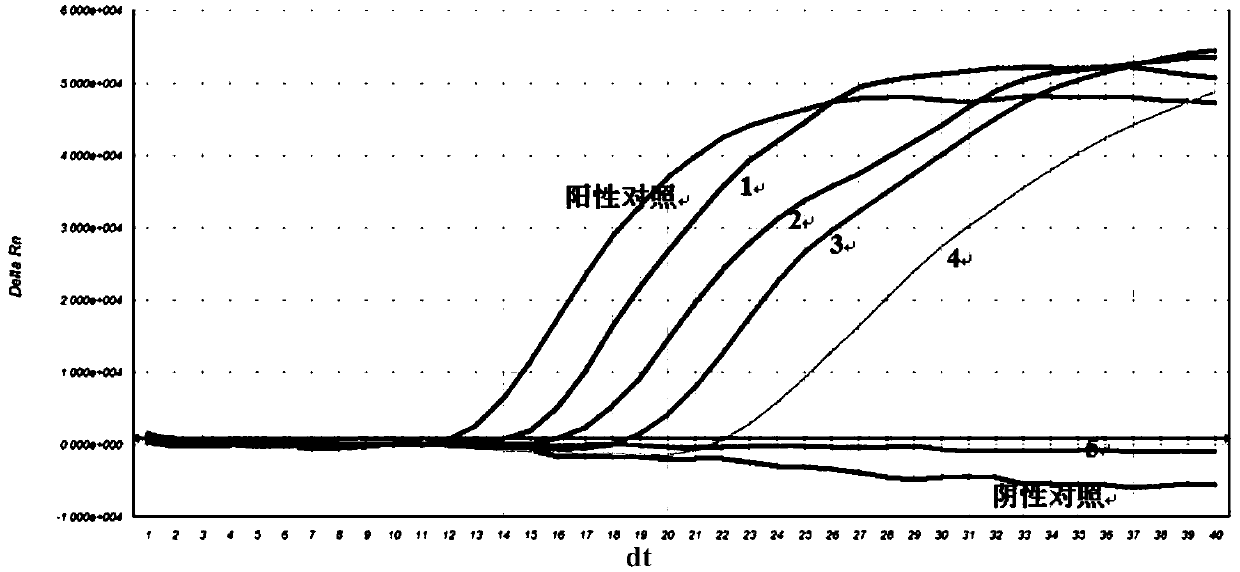
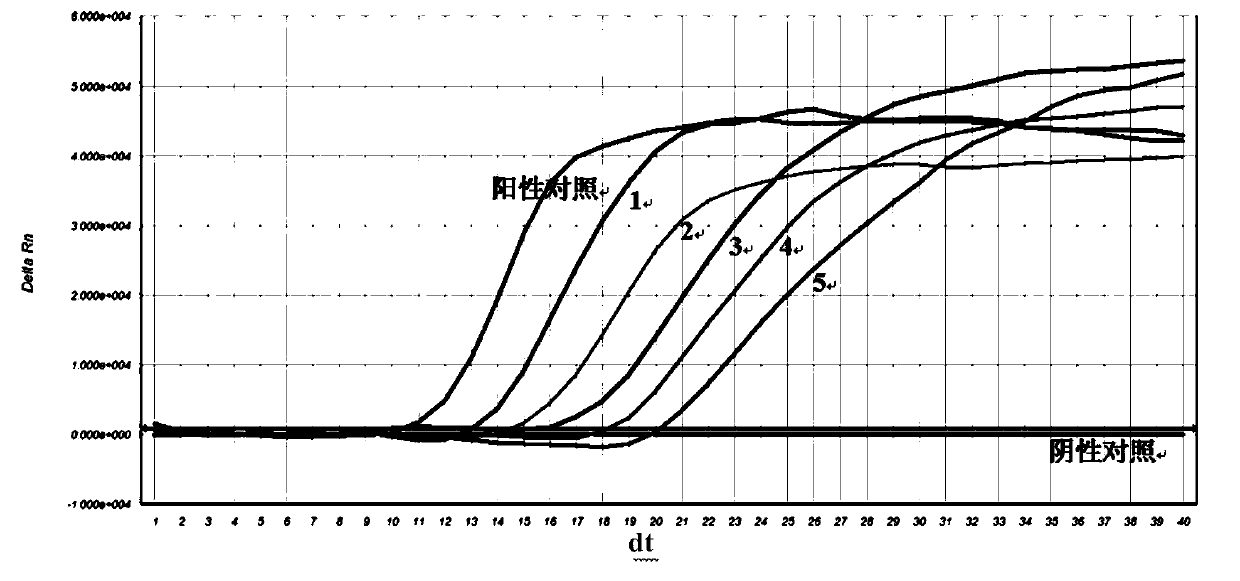
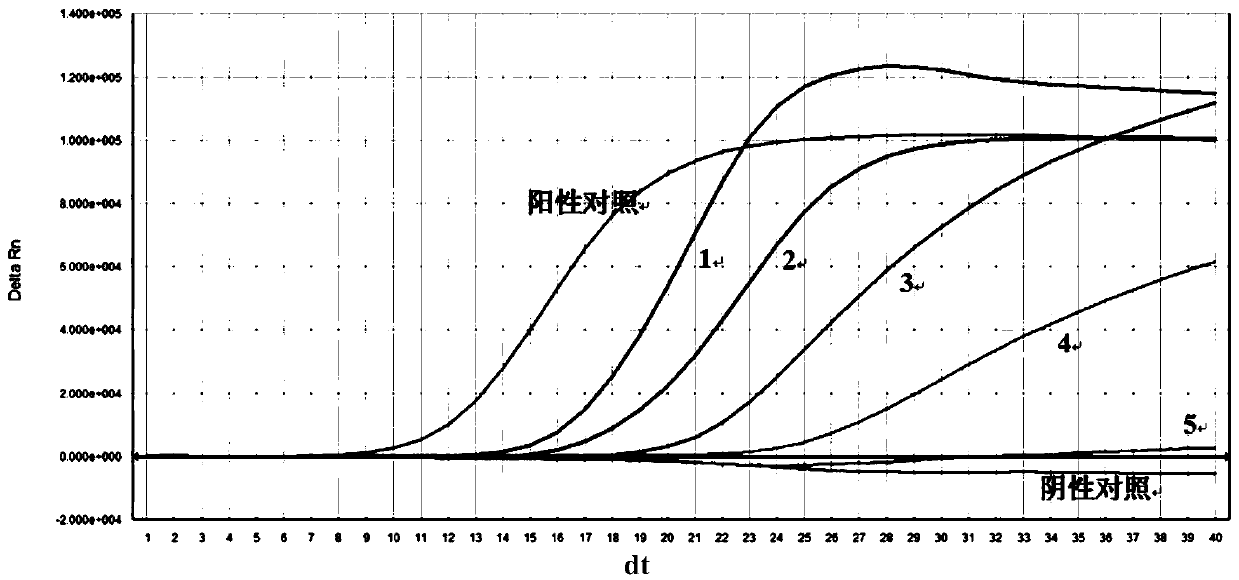
Nucleic acid constant-temperature amplification method of Methicillin-resistant Staphylococcus aureus (MRSA)
A methicillin, staphylococcus-resistant technology, applied in the field of bacterial biological detection, can solve problems such as no research reports yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0185] Example 1 Design of special primers and probes for real-time fluorescent nucleic acid constant temperature amplification detection of methicillin-resistant Staphylococcus aureus (MRSA)
[0186] The present invention selects highly conserved and specific segments in the MRSA (SA 23s rRNA and mecA) gene as the amplified target sequence region (the nucleotide sequences thereof are shown in SEQ ID No.: 1, SEQ ID No.: 2 ), according to the principle of primer probe design, using DNAStar, DNAMAN software and manual design for real-time fluorescent nucleic acid constant temperature amplification detection of methicillin-resistant Staphylococcus aureus (SA 23s rRNA and mecA of MRSA) special primers and probe sequences , to obtain the following specific sequence:
[0187] (1) A capture probe (TCO, Target Capture Oligo) that can specifically bind to the target nucleic acid (SA 23s rRNA) sequence of methicillin-resistant Staphylococcus aureus (MRSA), the nucleotide sequence of the...
Embodiment 2
[0214] Example 2 Preparation of a real-time fluorescent nucleic acid constant temperature amplification detection kit for methicillin-resistant Staphylococcus aureus (MRSA)
[0215] Using the special primers and probes provided in Example 1, the real-time fluorescent nucleic acid constant temperature amplification detection kit for methicillin-resistant Staphylococcus aureus (MRSA) of the present invention was obtained. The kit contains capture probe (TCO, TargetCapture Oligo), T7 primer, nT7 primer, MRSA (SA 23srRNA) detection probe, SA internal standard detection probe, MRSA (mecA) detection probe, mecA internal standard detection probe Needle, internal standard, M-MLV reverse transcriptase and T7 RNA polymerase and other components.
[0216] The capture probe is present in the nucleic acid extraction solution, and the T7 primer, nT7 primer, MRSA detection probe, and internal standard detection probe are respectively present in the MRSA (SA 23srRNA) detection solution and th...
Embodiment 3
[0244] Example 3 Methicillin-resistant Staphylococcus aureus real-time fluorescence nucleic acid constant temperature amplification detection method
[0245] 1. Sample collection, transportation and storage
[0246] sample collection
[0247] 1.1 Collection of nasal and oropharyngeal swab specimens: Take nasal and oropharyngeal secretions with medical cotton swabs, soak the swab head in 2mL of normal saline and stick to the tube wall to squeeze dry, add 2mL of sample preservation solution to it for later use, The sample is the sample to be tested.
[0248] 1.2 Wound secretion sample collection: Take the wound secretion with a medical cotton swab, soak the swab head in 2mL of normal saline and stick it to the wall of the tube to squeeze dry, add 2mL of sample preservation solution to it for later use, and this sample is the sample to be tested.
[0249] Specimen Transport and Storage
[0250] The collected specimens can be used for testing immediately or stored at -70°C for ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com