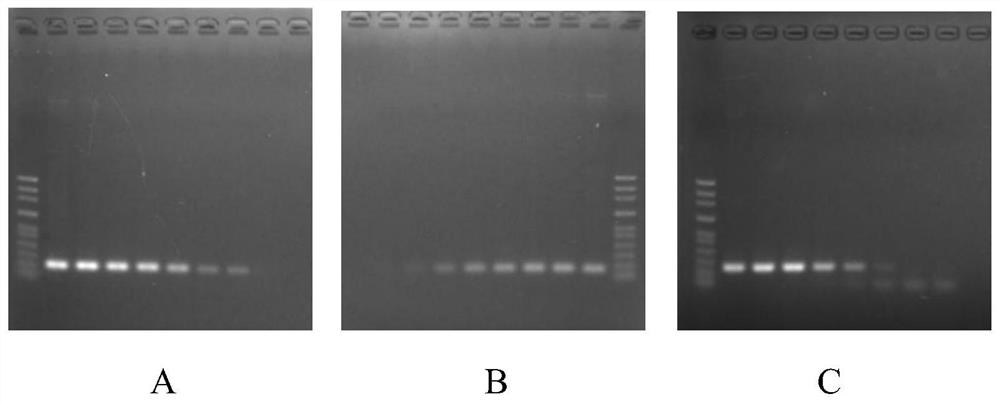
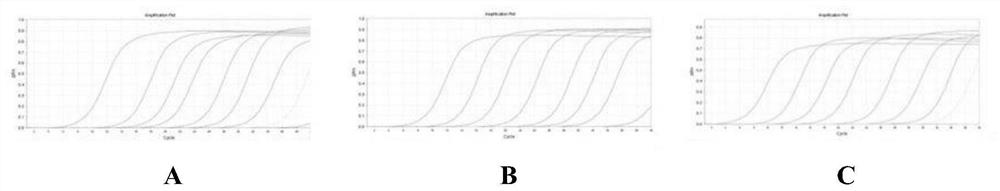
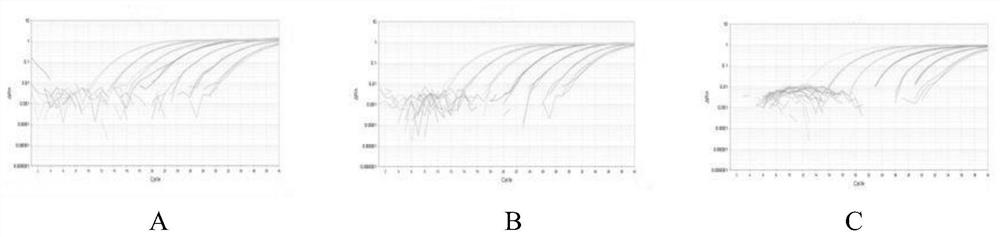
A triple fluorescent quantitative PCR detection method and kit for Rhizoctonia solani of cruciferous vegetables
A technology of cruciferous vegetables and Rhizoctonia solani, applied in the field of molecular biology, can solve problems such as difficult to distinguish, time-consuming disease detection, laborious fungal compound infection, etc., and achieve simple operation, shortened detection time, and high sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1 Synthesis of primers and probes
[0059] Synthesis of primers and probes for triple fluorescence quantitative PCR detection of R. solani fusion groups AG-2-1, AG-1-IB and AG-4HGII, the specific steps are as follows:
[0060] (1) Primers and probes of R. solani fusion group AG-2-1:
[0061] AG-2-1 upstream primer: 5'-GCTACAACCCCAAGTCGG-3' (SEQ ID NO: 1)
[0062] AG-2-1 downstream primer: 5'-GTAAAAGCGTAAAACCTGGGTG-3' (SEQ ID NO: 2)
[0063] AG-2-1 probe: 5'-FAM-CCTATCTCTGGATGGCACGGTGAC-TAMRA-3' (SEQ ID NO:3);
[0064] (2) Primers and probes of R. solani fusion group AG-1-IB:
[0065] AG-1-IB upstream primer: 5'-AACAAGATGGACACTACCAAGG-3' (SEQ ID NO:4)
[0066] AG-1-IB downstream primer: 5'-GGTCCTCGCTCCACTATAAAG-3' (SEQ ID NO:5)
[0067] AG-1-IB probe: 5'-VIC-AGTACAAACTCCACTCGGGTAACGACA-TAMRA3' (SEQ ID NO: 6);
[0068] (3) Primers and probes of R. solani fusion group AG-4HGII:
[0069] (1) AG-4HGII upstream primer: GCGCGGTAAAGGAATTTTGC (SEQ ID NO: 7)
[00...
Embodiment 2 3
[0072] Example 2 Preparation of triple fluorescent quantitative PCR detection premix
[0073] The preparation of triple fluorescent quantitative PCR detection mixture, the specific steps are as follows:
[0074] The following components synthesized in Example 1 were mixed well to obtain a mixed solution for the detection of R. solani fusion groups AG-2-1, AG-1-IB and AG-4HGII by triple fluorescence quantitative PCR.
[0075] AG-2-1 upstream primer: 0.2μL / test;
[0076] AG-2-1 downstream primer: 0.2μL / test;
[0077] AG-2-1 probe: 1μL / test;
[0078] AG-1-IB upstream primer: 0.3μL / test;
[0079] AG-1-IB downstream primer: 0.3μL / test;
[0080] AG-1-IB probe: 1μL / test;
[0081] AG-4HGII upstream primer: 0.2μL / test;
[0082] AG-4HGII downstream primer: 0.2μL / test;
[0083] AG-4HGII probe: 1 μL / test;
[0084] qPCR MIX: 10.4 μL / test;
[0085] Double distilled water: 4.2 μL / test.
Embodiment 3
[0086] The preparation of embodiment 3 positive control substance
[0087] Preparation of positive controls in the triple fluorescent quantitative PCR detection kit for R. solani fusion groups AG-2-1, AG-1-IB and AG-4HGII:
[0088] (1) Construction of plasmid
[0089] 1. Construction of the R. solani fusion group AG-2-1 plasmid: PCR was used to amplify the nucleic acid of the R. solani fusion group AG-2-1 sample (preserved by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences). , the obtained sequence is:
[0090] 5'-GCTACAACCCCAAGTCGGTCGCTTTCGTCCCTATTTCTGGATGGC ACGGTGACAACATGTTGGAGGAGTCCGTCAAGTACGTCATACTGTCGCACCCAGGTTTACGCTTTTAC-3' (SEQ ID NO: 10) nucleic acid fragment of the target amplification sequence, the primer sequences are shown in SEQ ID NO: 1-3. After purification of the amplified fragment, it was cloned into pMD19-T vector by TA, and the recombinant vector with correct sequencing result was transformed into DH5α and amplified to obt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com