Metagenome sequencing quality control prediction evaluation method and model
A technology of metagenomics and evaluation modules, which is applied in the field of metagenomics sequencing quality control prediction and evaluation methods and models, and can solve problems such as not being fully applicable, unable to calculate theoretical detection performance, and lacking theoretical support.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] A metagenomic sequencing quality control prediction evaluation method, comprising the following steps:
[0108] 1. Q20 Threshold System Evaluation Process.
[0109] Construct a Q20 threshold model, obtain sequencing parameters in the scheduled sequencing process, input the sequencing parameters into the Q20 threshold model for solution, and obtain the relationship between the ratio of Q20 and the correct rate.
[0110] The specific process is as follows:
[0111] 1. Get parameters
[0112] 1) Load the model calculation function to the R environment.
[0113]source("Q20.r")
[0114] 2) According to the scheduled sequencing process, set the parameters (values corresponding to the parameters are set according to actual requirements), run the loaded function, and obtain the result.
[0115] In this embodiment, parameter 1: the amount of sequencing data is set to 20M (a=20)
[0116] Parameter 2: The length of the sequencing fragment is set to 50bp (b=50)
[0117] Par...
Embodiment 2
[0187] A metagenomic sequencing quality control prediction evaluation model, including:
[0188] Data input module: used to obtain sequencing parameters, data parameters and strain mutation parameters in the scheduled sequencing process;
[0189] Model calculation module: perform evaluation and analysis according to the metagenomic sequencing quality control prediction evaluation method in Example 1;
[0190] Result output module: used to output the evaluation and analysis results of the model calculation module.
Embodiment 3
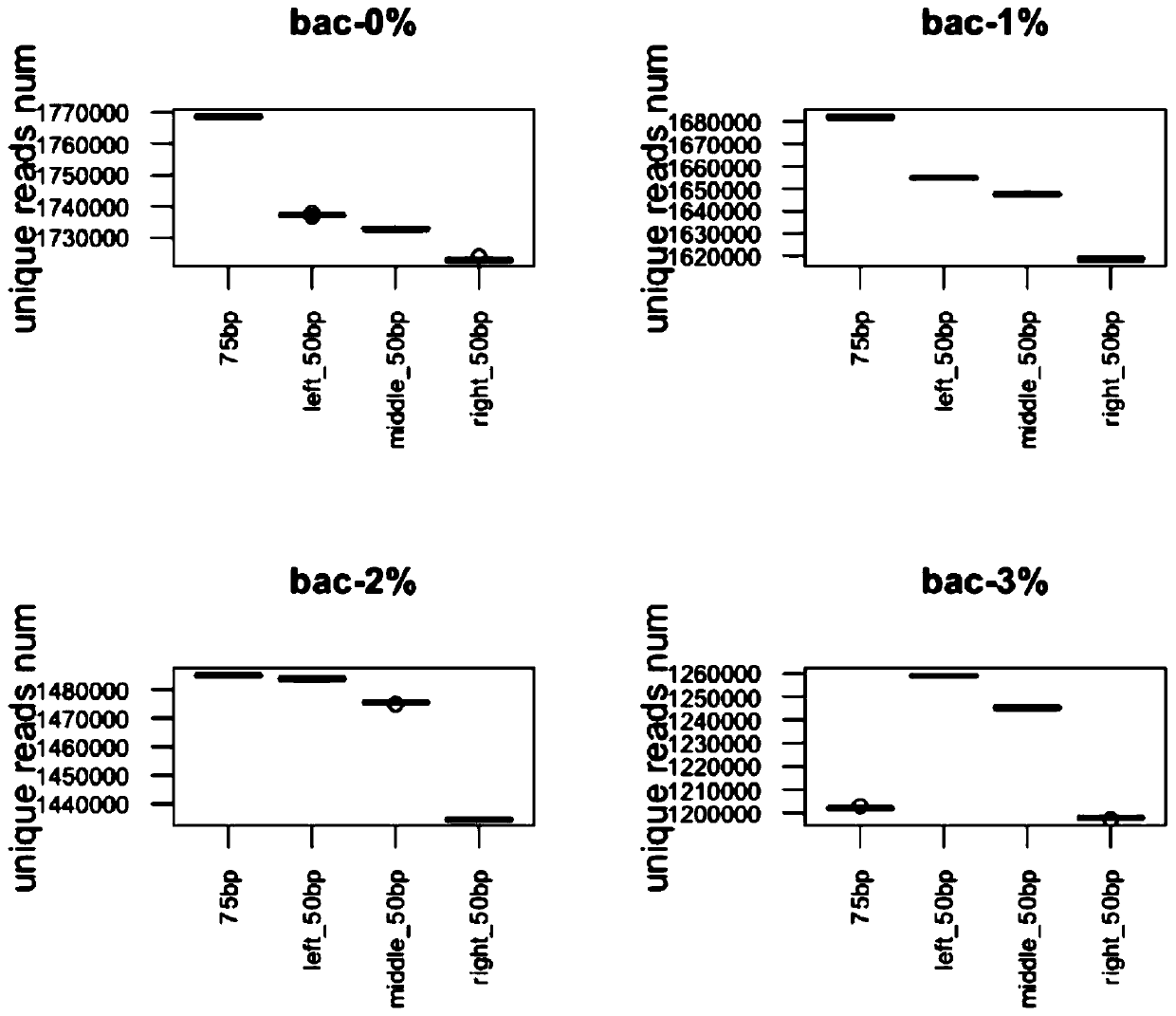
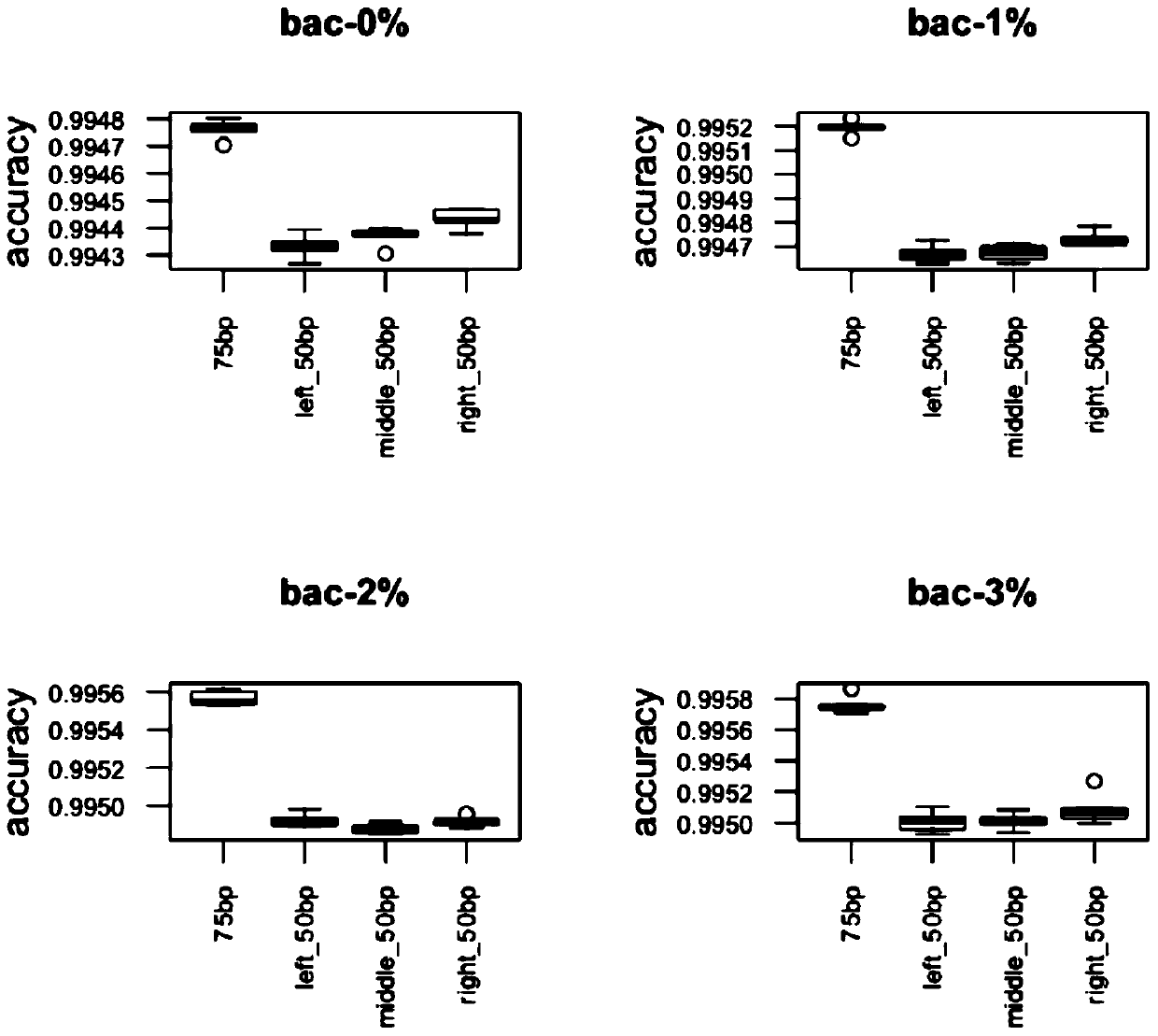
[0192] Verification test analysis and comparison.
[0193] 1. Comparison of Q20 threshold system.
[0194] Dilute Streptococcus agalactiae to 6 different concentrations with primary water: 10cfu / ml, 10 2 cfu / ml,10 3 cfu / ml, 10 4 cfu / ml,10 5 cfu / ml,10 6 cfu / ml, 6 samples were obtained.
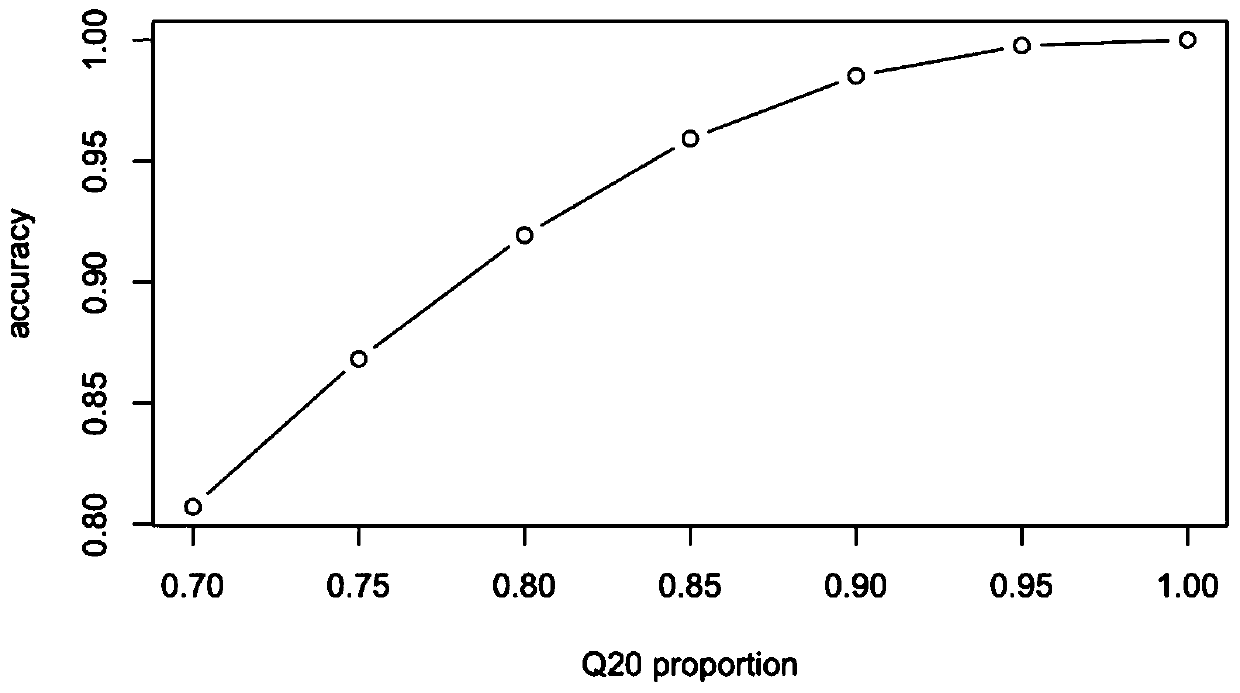
[0195] Carry out NGS sequencing and data analysis according to the predetermined sequencing process in Example 1, and calculate the relationship between the Q20 ratio and "correct rate" of the 6 samples, and compare the theoretical data, the results are as follows Figure 10 shown.
[0196] It can be seen from the figure that, considering that there is a certain proportion of sudden changes in the actual data, the theoretical value obtained from the above analysis is basically consistent with the actual performance.
[0197] 2. Comparison of sequencing data volume threshold systems.
[0198] Continuing to use the above Streptococcus agalactiae for comparison, the main parameters of this...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com