Methylation detection kit for nasopharyngeal carcinoma related genes DACT1, NFAT1 and SHISA3
A detection kit and technology for nasopharyngeal carcinoma, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve problems such as non-specific amplification, PCR pollution, etc., to improve specificity, eliminate PCR pollution, improve The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1: Preparation of kits for detection of methylation of nasopharyngeal carcinoma-related genes DACT1, NFAT1 and SHISA3
[0052] Kits for detection of methylation of nasopharyngeal carcinoma-related genes DACT1, NFAT1 and SHISA3, including fluorescent PCR reaction system, negative quality control and positive quality control. The preparation of the kit includes the following steps:
[0053] (1) Preparation of alkaline solution: Take an appropriate amount of NaOH to prepare a solution with a concentration of 75 mM and a pH value of 12.5, which is an alkaline solution, and save it for future use.
[0054] (2) Design and preparation of primers and probes: Design several sets of primers and probes for the DACT1, NFAT1, and SHISA3 gene CpG islands and ACTB genes, respectively, and conduct pre-experiments on each primer and probe to compare sensitivity, specificity and other properties Finally, four sets of LNA modified primers and specially designed competitive fluore...
Embodiment 2
[0070] Example 2: Validation experiment of fluorescent PCR reaction system for detection of non-desulfurized samples
[0071] (1) Purpose of the experiment
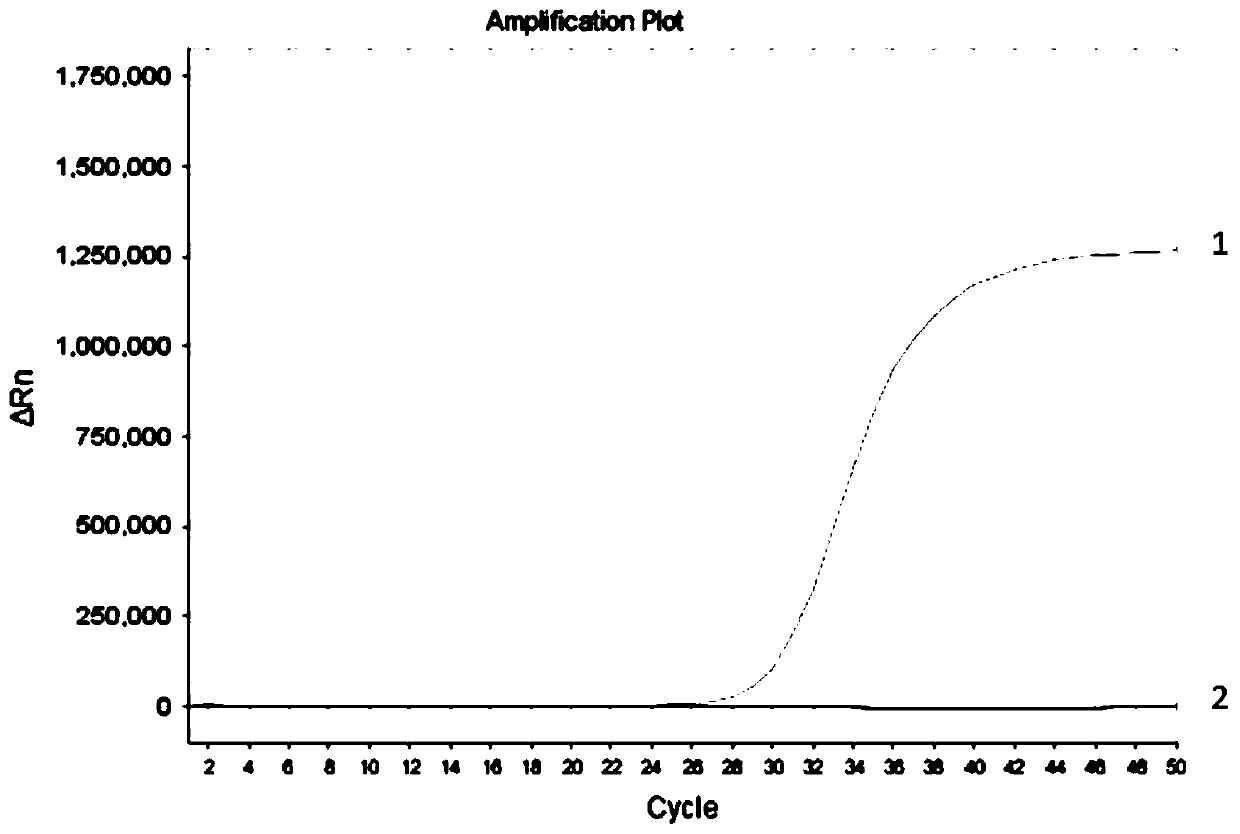
[0072] In this example, by comparing the detection results of the desulfurized samples and non-desulfurized samples with the conventional fluorescent PCR system and the fluorescent PCR system of the present invention without UDG enzyme, to verify the effectiveness of the fluorescent PCR reaction system of the present invention in the detection of non-desulfurized samples sex.
[0073] (2) Experimental method
[0074] In this example, methylated human genomic DNA was used, and after bisulfite conversion treatment, it was divided into two groups, one of which was desulfurized by NaOH and purified to obtain desulfurized DNA samples, and the other group was directly desulfurized without desulfurization. Purified, that is, non-desulfurized DNA samples. The above-mentioned two groups of DNA samples were used as templates, re...
Embodiment 3
[0086] Embodiment 3: Effect Analysis Experiment of Fluorescent PCR Reaction System Eliminating PCR Contamination
[0087] (1) Purpose of the experiment
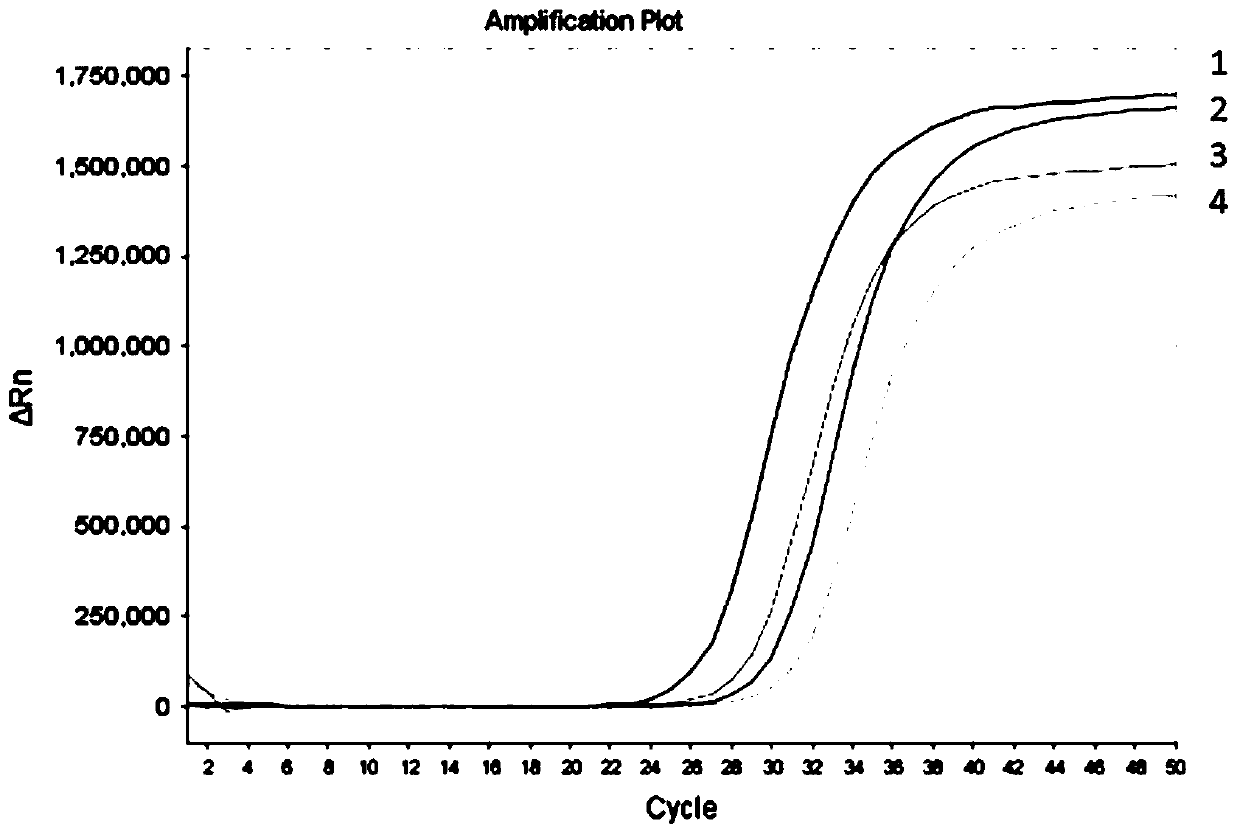
[0088] In this example, the effect of the fluorescent PCR reaction system of the present invention on eliminating PCR contamination was analyzed by comparing the detection results of samples containing or not containing PCR pollutants with the fluorescent PCR system of the present invention without UDG enzyme.
[0089] (2) Experimental method
[0090] In this example, non-methylated human genomic DNA that has been converted by bisulfite but not desulfurized, methylated human genomic DNA that has been converted by bisulfite but not desulfurized, and PCR pollutants are selected to prepare 4 kinds Samples with or without PCR contamination. Among them, PCR pollutants are amplification products of methylated human genomic DNA. See the table below for the preparation of the 4 samples with or without PCR contaminants.
[0091] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com