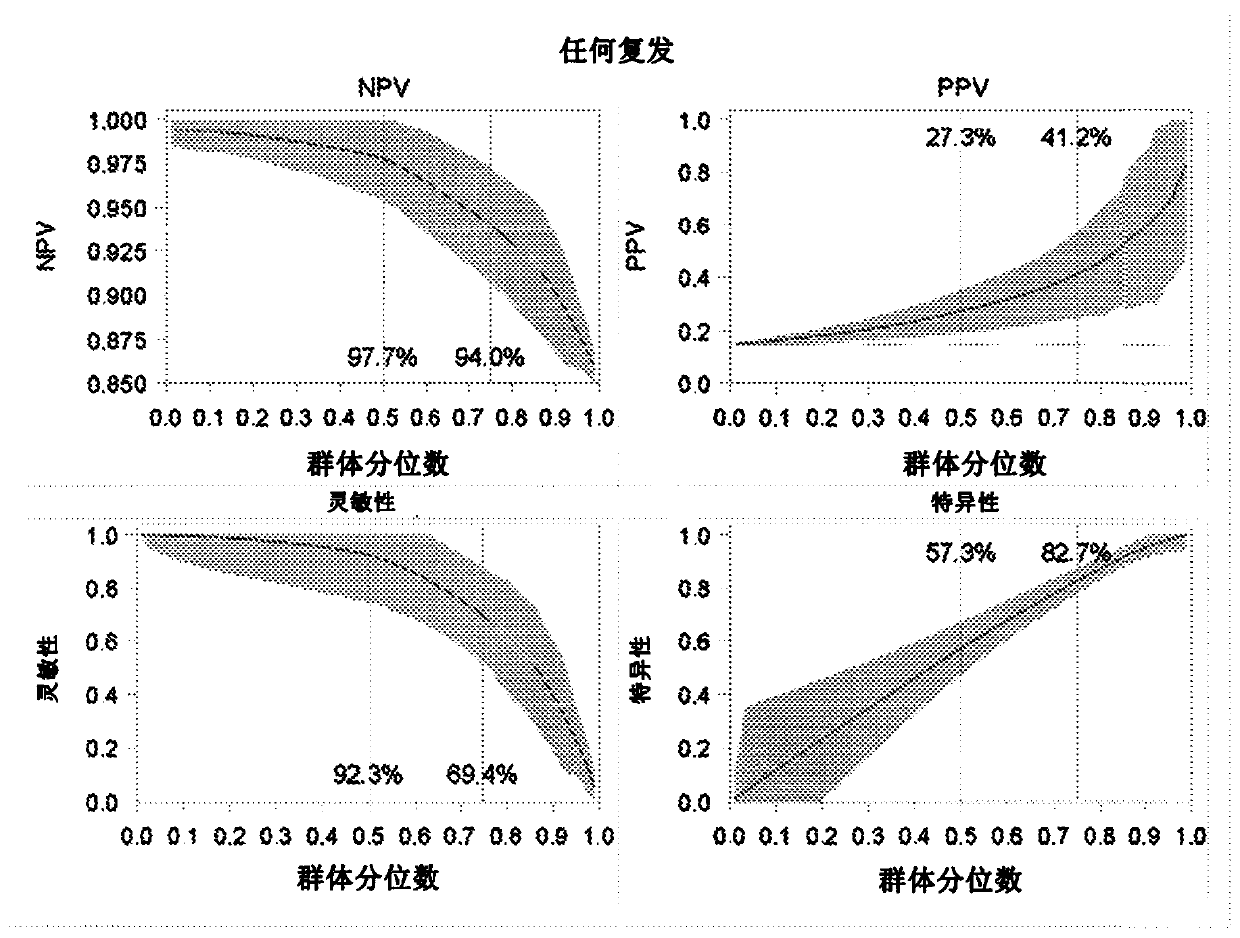
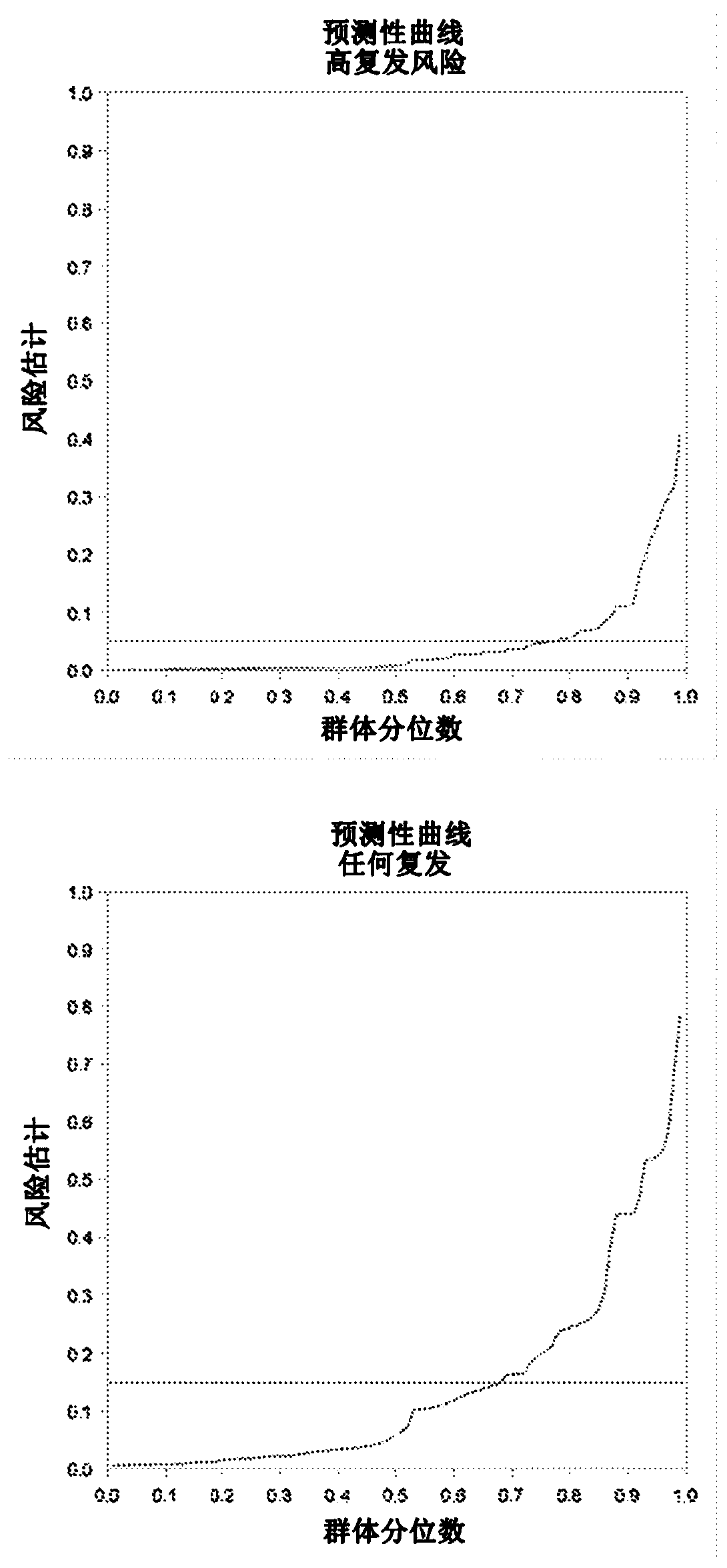
DNA methylation and mutational analysis methods for bladder cancer surveillance
A technology of bladder cancer and methylation, applied in the fields of biochemical equipment and methods, analytical materials, biomaterial analysis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0132] Example 1: Method of evaluating methylation and proof-of-concept study
[0133] Urine cell sediments from 66 patients with bladder cancer (25 patients without recurrence, 14 low-risk (LR) (Ta low grade) patients, and 27 high-risk (HR) (T1 or high grade) patients) were used for conception Validation (POC) study. 11 methylation gene markers and 1 methylation-independent (MIP) reference gene selected based on literature were used to assess recurrence. Methylation-specific quantitative PCR (MS-qPCR) analysis after bisulfate conversion was used to determine the methylation levels of these 12 genes. This study was used to evaluate the performance of individual methylation markers and the combined score for predicting the presence of recurrence. Based on this research, several methods for evaluating methylation signals have been developed and used in the refinement of a larger set of candidate markers.
[0134] The methylation fraction (MF) of each gene is estimated from the met...
Embodiment 2
[0143] Example 2: Methylome discovery study
[0144] A gene discovery study was conducted to identify other gene candidates that are differentially methylated in samples from patients with bladder cancer and patients without cancer. A total of 26 tumor tissue samples and 7 urine cells from cancer patients (14 patients previously diagnosed with low risk of recurrence (LR) and 19 patients previously diagnosed with high risk of recurrence (HR)) were used in the analysis The sediment, as well as 7 urine cell sediments and 5 tissue samples from cancer-free patients (7 healthy individuals and 5 patients who were not previously diagnosed with bladder cancer or who were previously diagnosed with no malignancy at the time of surgery). Based on the analysis of the Cancer Genome Atlas (TCGA), the methylome assessment targets approximately 6 million nucleotides in regions that are differentially methylated in 11 cancers, and approximately 724,000 sites are sequenced for each sample. The fol...
Embodiment 3
[0146] Example 3: Refinement study
[0147] This detailed study evaluated 96 markers on 203 previously diagnosed bladder cancer patients currently under surveillance, including 86 methylation marker genes from gene discovery studies, 4 methylation-independent reference genes, and 6 FGFR3 mutations. Two of the six FGFR3 mutation assays failed to generate any signal, and further evaluation of these two was excluded. In addition, 170 patients in this study had enough RNA for other evaluations, including the determination of 2 TERT mutations. The results presented here are based on these 170 patients, which evaluated 86 methylation markers, 4 reference genes, 4 FGFR3 mutations, and 2 TERT mutations. The patients included 85 patients who had not relapsed, 32 patients who were previously diagnosed with LR relapse, and 53 patients who were previously diagnosed with HR relapse.
[0148] After bisulfate conversion, RT-PCR analysis was used to determine the methylation level. Non-paramet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com