Glucose dehydrogenase mutant and preparation method thereof
A glucose dehydrogenase and mutant technology, applied in the field of glucose dehydrogenase mutants and its preparation, can solve problems such as difficulties in expression and separation and purification, poor substrate specificity, and restrictions on the application of blood glucose measurement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1. Construction of recombinant plasmid pPIC9K-fad-gdh
[0052] Using the restriction-free (RF) cloning method, 9K99a F and 9K99a R were used as primers, and fad-gdh (SEQ ID NO: 2) was used as a template for PCR amplification. Use the SanPrep Column PCR Product Purification Kit (Sangon Bioengineering (Shanghai) Co., Ltd.) to purify the PCR amplified product, use the purified product as a primer, use the pPIC9K plasmid as a template, and use Takara PrimeSTAR GXL DNA polymerase In the second PCR, the recombinant plasmid pPIC9K-fad-gdh was obtained.
[0053] Among them, the primer sequence is:
[0054] 9K99a F:
[0055] 5-CTCTCGAGAAAAGAGAGGCTGAAGCTTCACACAGGAAACAGACCATG-3' (SEQ ID NO: 3),
[0056] 9K99aR:
[0057] 5-GGAACAGTCATGTCTAAGGCGAATTACATCCGCCAAAACAGCCAAGC-3' (SEQ ID NO: 4).
Embodiment 2
[0058] Embodiment 2. Obtain the mutant construct of the Y406 site of fad-gdh
[0059] Using the random mutation primer Y406X-F / R at the Y406 site, the recombinant plasmid pPIC9K-fad-gdh prepared in Example 1 was used as a template to perform site-directed saturation mutation. The saturation mutant PCR products were transformed into XL10-GOLD competent cells for cloning. Plasmids were extracted using an extraction kit (Sangon Bioengineering (Shanghai) Co., Ltd.), and the plasmids were sequenced. From it, 19 kinds of plasmid pPIC9K-fad-gdh with mutation at Y406 site of FAD-GDH were screened.
[0060] Among them, the random mutation primers are:
[0061] Y406X-F:
[0062] 5'-TGCCGAAGTTTTAAAANNNCCAGGCTCCGCCAC-3' (SEQ ID NO: 5)
[0063] Y406X-R:
[0064] 5'-GTGGCGGAGCCTGGNNNGTTTAAAACTTCGGCA-3' (SEQ ID NO: 6).
Embodiment 3
[0065] Example 3. Expression of FAD-GDH in recombinant Pichia strains
[0066] The mutant plasmid prepared in Example 2 was linearized, and then the mutant plasmid was transformed into Pichia pastoris cells using a gene transfer instrument.
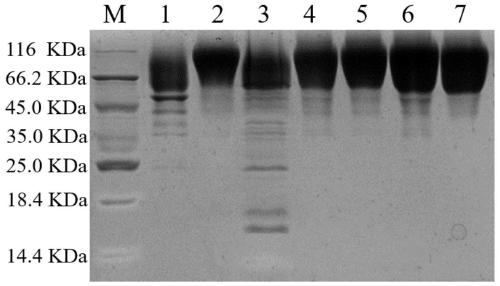
[0067] Spread the recombinant Pichia pastoris on the MD plate and incubate at 30°C for 2-3 days until white colonies grow. Use a toothpick to pick the white single colony on the MD plate, inoculate it on a YPD plate containing 0.5 mg / mL geneticin, and incubate at 30°C for 2 to 3 days until the white colony grows. Use a toothpick to pick up the white single colony grown on the YPD plate containing 0.5 mg / mL geneticin, inoculate it into the YPD plate containing 1.0 mg / mL geneticin, and culture it at 30°C for 2-3 days until the white colony is formed. Taking Y406E (the 406th tyrosine is replaced by glutamic acid) as an example, the result is as follows figure 1 As shown in A~1D, figure 1 The results of screening for multi-copy recombinant...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com