Research method of Tet2 sequence preference
A preference and sequence technology, applied in the research field of Tet2 sequence preference, can solve problems such as the influence of antibody affinity and specificity, subtle changes cannot provide effective quantitative analysis data, etc., and achieve low detection cost, simple operation, and biological good compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Tet2 pair sequence binding preference analysis
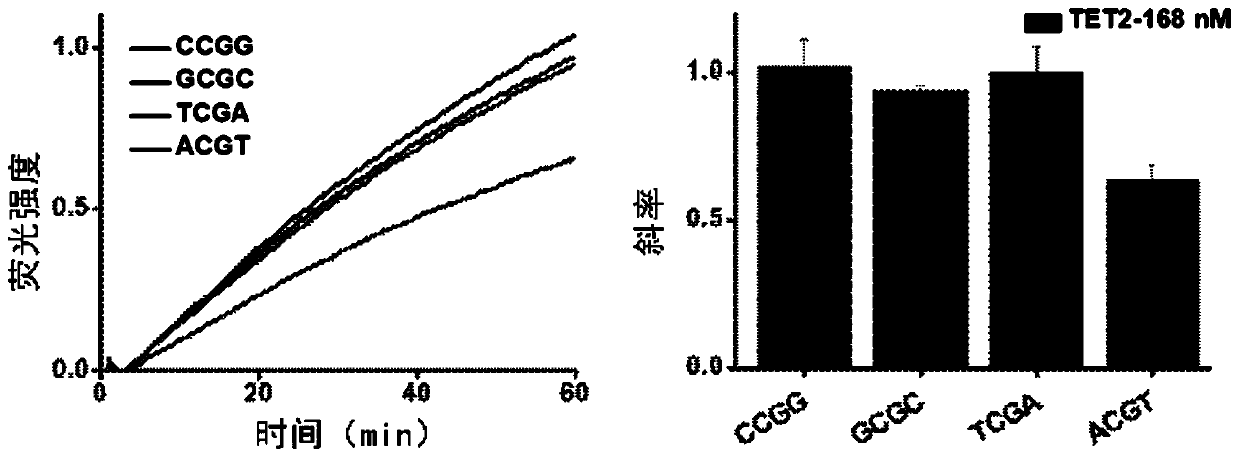
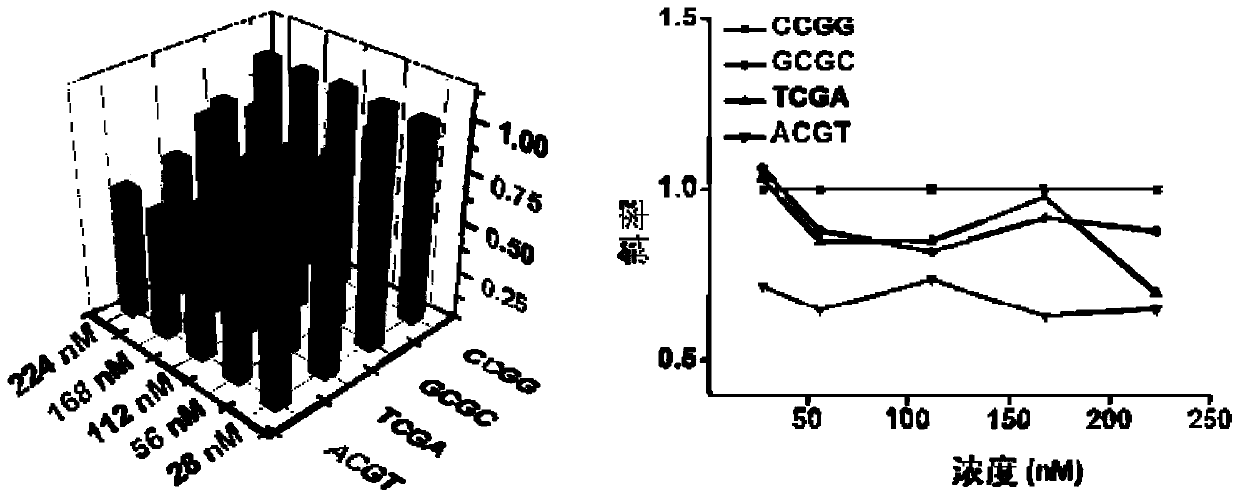
[0054] The binding preference of Tet2 to the four most commonly methylated DNA sequences was investigated. Based on Tet2's different preference for the flanking position of the action site, different sequences bind different amounts of protein, which leads to different extension rates of DNA polymerases and differences in fluorescence amplification curves during the sequence amplification process.
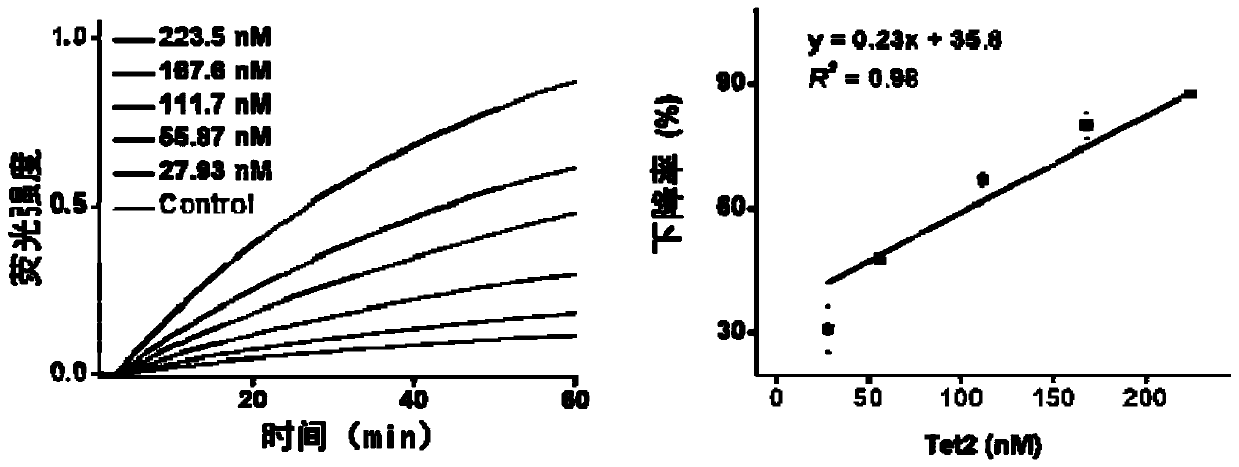
[0055] The effect of Tet2 concentration on the sequence amplification reaction from low to high, respectively 27.9, 55.9, 112, 168 and 224nM was studied.
[0056] (1) Isothermal amplification reaction to detect the effect of Tet2 on different sequences
[0057] 10mM HEPE solution, 200nM Fe 2 SO 4 Solution, 4mM ascorbic acid, 2mMα-ketoglutarate, 150mMNaCl, 1mM ATP, 168nM Tet2 protein, continue to add 1×Cutsmart buffer (20mM Tris-acetate, 50mM potassium acetate, 10mM magnesium acetate, 0.1mg mL –1 BSA, pH 7.9), 125 μM dNTPs mi...
Embodiment 2
[0063] Study on the Oxidation Preference of Tet2 to Different Sequences
[0064] Carry out glycosylation modification on the DNA sequence after Tet2 oxidation, and further use 3-carboxyphenylboronic acid (3-CPBA) to condense with the cis-ortho-diol on the sugar ring to obtain the modified site CPBA-5gmC, which will contain 3CPBA The sequence of the -gmC site and the unmodified DNA sequence were amplified separately, and the amplification efficiency of the two was compared to determine the oxidation preference of Tet2.
[0065] The amplification reaction system includes two parts, A and B, wherein part A contains nickase buffer, dNTPs, primers and templates, and part B contains polymerase buffer, nucleic acid dye, polymerase and nickase. Optimize the amplification reaction conditions first.
[0066] (a) template usage
[0067] Prepare template solutions with different concentrations to optimize the optimal template dosage. The reaction is divided into two parts, Part A and P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com