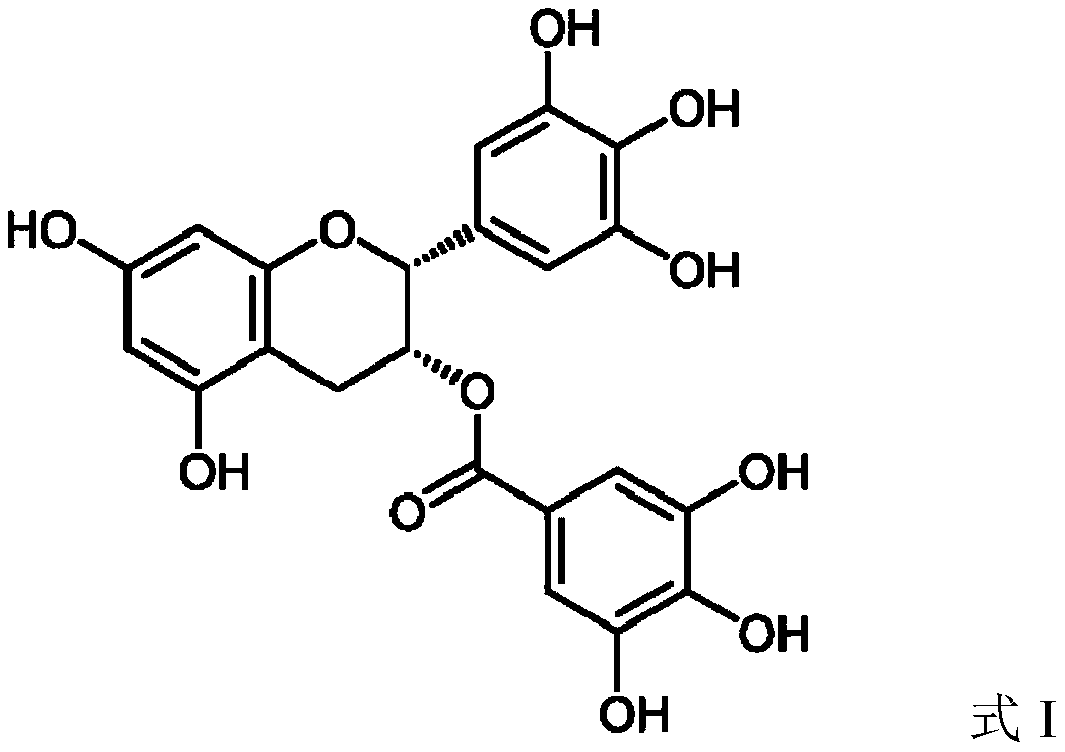
Method for methylating EGCG through enzymatic catalysis
An enzymatic catalysis and methylation technology, applied in microorganism-based methods, biochemical equipment and methods, enzymes, etc., can solve the problems of low enzyme catalysis efficiency, difficult to meet the needs of actual production, etc., and achieve efficient methylation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Construction of recombinant escherichia coli
[0032] Using a high-efficiency plant genomic DNA extraction kit (purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd.), the genome was extracted from commercially available Benifuuki tea leaves according to the manufacturer's instructions, and then, the genome was extracted using a method purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd. TIANSeq high-fidelity PCR reaction master mix, according to the manufacturer's instructions with primers F and R (F: CATATGGCGACCAACGGTGAA (SEQ ID NO.: 1); R: CTCGAGGCAAACACG (SEQ ID NO.: 2)) for PCR cloning (PCR program Pre-denaturation at 94°C for 5 minutes; 94°C for 30s, 50°C for 50s, 72°C for 1min, 35 cycles; 72°C for 10min), use 1.0% agarose gel electrophoresis to detect PCR amplification products, PCR amplification products according to the purchased The nucleic acid sequence encoding O-methyltransferase (SEQ ID NO.: 3; referred to as "O-meth...
Embodiment 2
[0034]Example 2: Cultivation of recombinant Escherichia coli and expression of O-methyltransferase
[0035] A single colony of the positive recombinant bacteria obtained in Example 1 was picked and inoculated into fresh LB medium, and cultured at 37° C. for 8 hours as a seed solution.
[0036] The above-mentioned seed liquid is inoculated into the fermentation medium according to the inoculum size of 5% (v / v) (glucose 20g / L, yeast extract powder 5g / L, potassium dihydrogen phosphate 1g / L, magnesium sulfate 0.2g / L, manganese sulfate 0.2g / L, and the remaining amount of water), through exponential feeding of glucose and yeast powder, the fermentation process of recombinant Escherichia coli is controlled, and the dissolved oxygen concentration is kept at 20%-30%. Grow to OD 600 At 60, the expression of O-methyltransferase was induced by adding 10% (w / v) lactose at a constant flow rate of 0.04 mL / min; the fermentation temperature was 30°C. Samples were taken at different time poin...
Embodiment 3
[0037] The preparation of embodiment 3 crude enzyme liquid
[0038] The fermentation broth obtained in Example 2 was centrifuged at 12000rpm for 10min, and the thalline was collected, and an equal volume of 20mM PBS buffer (20mmol / L NaH 2 PO 3 , 20mmol / L Na 2 HPO 3 , 1mmol / L DTT, pH 7.4) to resuspend the bacteria (dilute according to the concentration of the bacteria to make the OD 600 10), sonicated (sonicated for 2 s, intermittently for 2 s, 5 min in total), and centrifuged at 12000 rpm for 10 min to collect the supernatant to obtain a crude enzyme solution.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com