A method for detecting (CAG) n repeat sequences using rnase H
A nucleotide sequence and kit technology, applied in the field of molecular biology, can solve the problems of non-reusable target DNA, difficult detection due to few fluorophores, poor detection accuracy, etc., and achieves enhanced detection sensitivity, high sensitivity, Detect fast effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
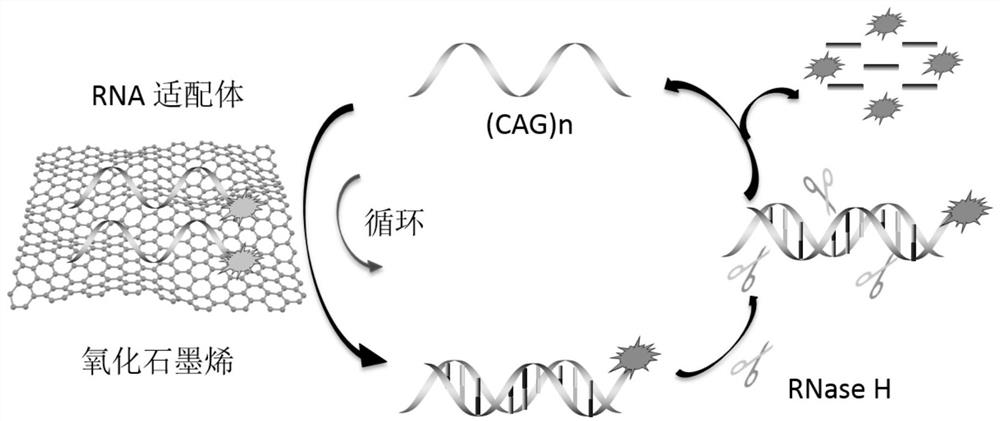
[0045] Working principle Feasibility analysis:
[0046] (1) Dilute graphene oxide to 100 μg / ml with DEPC water, mix well, and set aside.
[0047] (2) Dissolve fluorescently labeled aptamer R1 (sequence 5'-FAM-GUCGUC GUC GUC GUCGUC GUC GUC GUC GUC GUC-3') with DEPC water, and prepare 20×10 -6 mol / L solution, mix well and set aside.
[0048] (3) Dissolve (CAG)n (sequence is 5'-CAGCAG CAG CAG CAG CAG CAG CAG CAG CAG-3') with DEPC water to prepare 100×10 -6 mol / L solution, mix well and set aside.
[0049] (4) Dilute the fluorescently labeled aptamer R1 solution prepared in step (2) with DEPC water to 200×10 -9 mol / L, mix well, and become a tube ①.
[0050] (5) Add the graphene oxide solution prepared in step (1) at a final concentration of 24 μg / ml into tube ①, mix well, and incubate at room temperature for 10 min to form tube ②.
[0051] (6) Add a final concentration of 25×10 to tube ② -9 mol / L (CAG)n prepared in step (3), mix well, incubate at room temperature for 30 min, ...
Embodiment 2
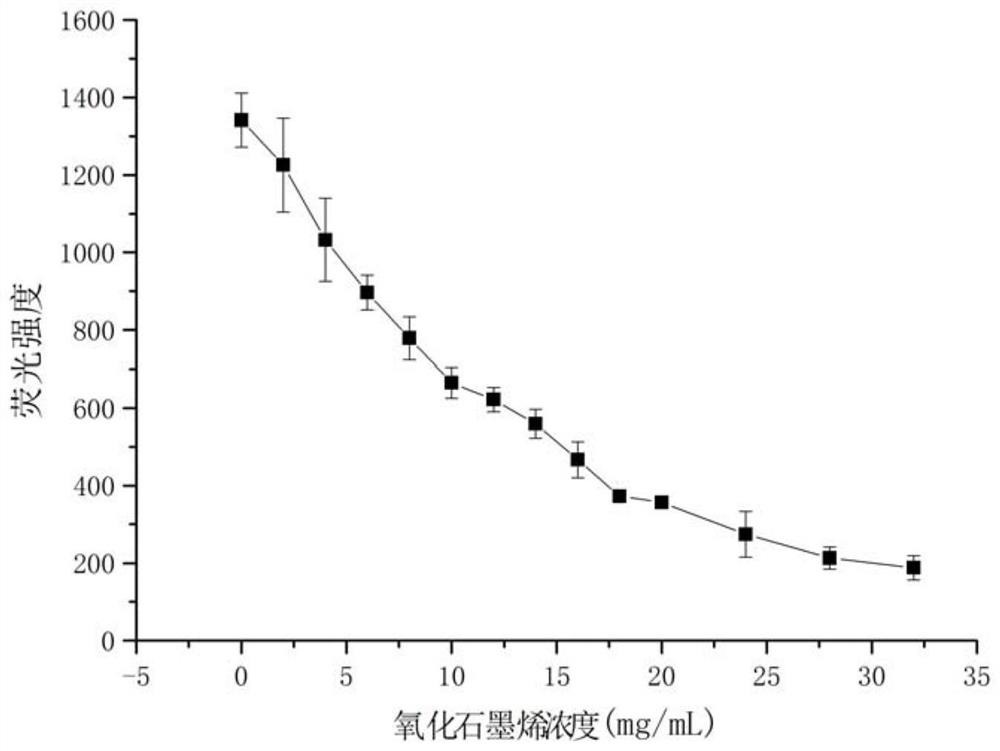
[0056] The ability of graphene oxide concentration to quench the fluorescence of aptamer RNA:
[0057] 1. Dilute graphene oxide to 100μg / ml with DEPC water and mix well.
[0058] 2. Dissolve fluorescently labeled aptamer R1 (sequence 5'-FAM-GUCGUC GUC GUC GUCGUC GUC GUC GUC GUC GUC-3') with DEPC water, and prepare 20×10 -6 mol / L solution, mix well.
[0059] 3. Add different final concentrations of graphene oxide solutions (0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 24, 28, 32 μg / ml) to the Tris-HCl buffer solution 20×10 in -6 mol / L FAM-R1, mix well, and incubate at room temperature for 30min.
[0060] 4. Use a Hitachi F-7000 fluorescence spectrophotometer. The excitation wavelength is 480nm, the excitation slit is 5nm, the emission slit is 2.5nm, the scanning speed is 1200nm / min, and the fluorescence intensity at 520nm is detected. The results showed that as the concentration of graphene oxide increased, the fluorescence intensity decreased continuously. After 24 μg / ml, the f...
Embodiment 3
[0063] To investigate the effect of the fluorescence quenching time of the aptamer, the specific steps are as follows:
[0064] 1. Dilute graphene oxide to 100μg / ml with DEPC water and mix well.
[0065] 2. Dissolve fluorescently labeled aptamer R1 (sequence 5'-FAM-GUCGUC GUC GUC GUCGUC GUC GUC GUC GUC GUC-3') with DEPC water, and prepare 20×10 -6 mol / L solution, mix well.
[0066] 3. Dissolve (CAG)n (sequence is 5'-CAGCAG CAG CAG CAG CAG CAG CAG CAG-3') with DEPC water, and make 100×10 -6 mol / L solution, mix well.
[0067] 4. In the containing 200×10 -9 Add 24 μg / ml graphene oxide solution to the mol / L Tris-HCl buffer solution of FAM-R1, mix well, and incubate at room temperature for 0, 2, 5, 8, 10, 12, 15, 20, 25, and 30 min, respectively.
[0068] 5. Use a Hitachi F-7000 fluorescence spectrophotometer. The excitation wavelength is 480nm, the excitation slit is 5nm, the emission slit is 2.5nm, the scanning speed is 1200nm / min, and the fluorescence intensity at 520nm is ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap