Method and device for haploid typing and variation detection of diploid genome sequencing fragments
A genome sequencing and diploid technology, applied in genomics, sequence analysis, proteomics, etc., can solve the problems of low sensitivity, short typing region length, poor accuracy, etc., and achieve high accuracy and high sensitivity typing The effect of improving the analysis and mutation detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0089] Capture and sequence the HLA target region on human chromosome 6, and analyze the information of the full-length region of the HLA-DRB1 gene.
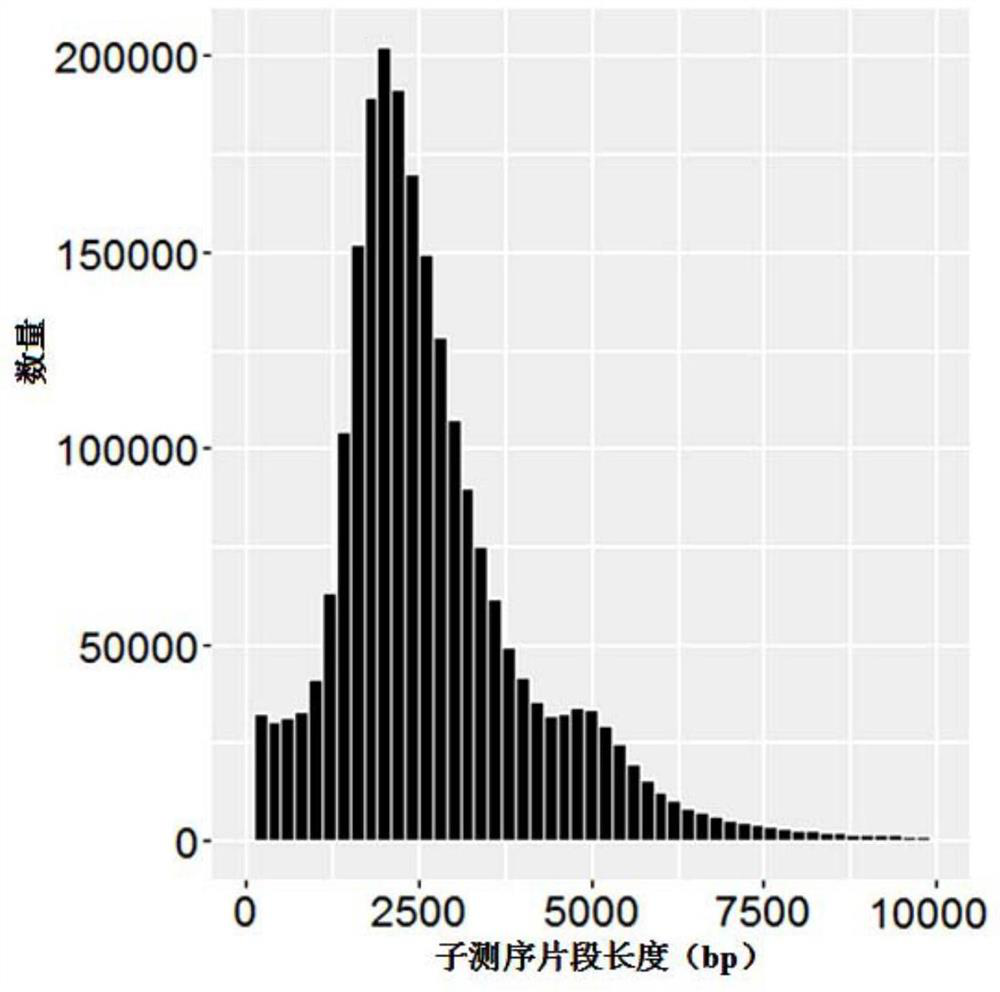
[0090] 1) The BGI-YH cell line sample is captured by the HLA chip using the mature and public experimental technology, that is, a DNA library with a length of 10k is constructed, and then sequenced using the PacBio RSII sequencer. And for the same BGI-YH sample, 5 parallel and independent capture, library construction, and sequencing operations were performed. The length distribution of sequenced fragments is as follows figure 2 As shown, the curve is smooth and contains the main peak of the sequenced fragment (subreads) with a length of 2.5k and the more obvious tailing around the length of 5k.
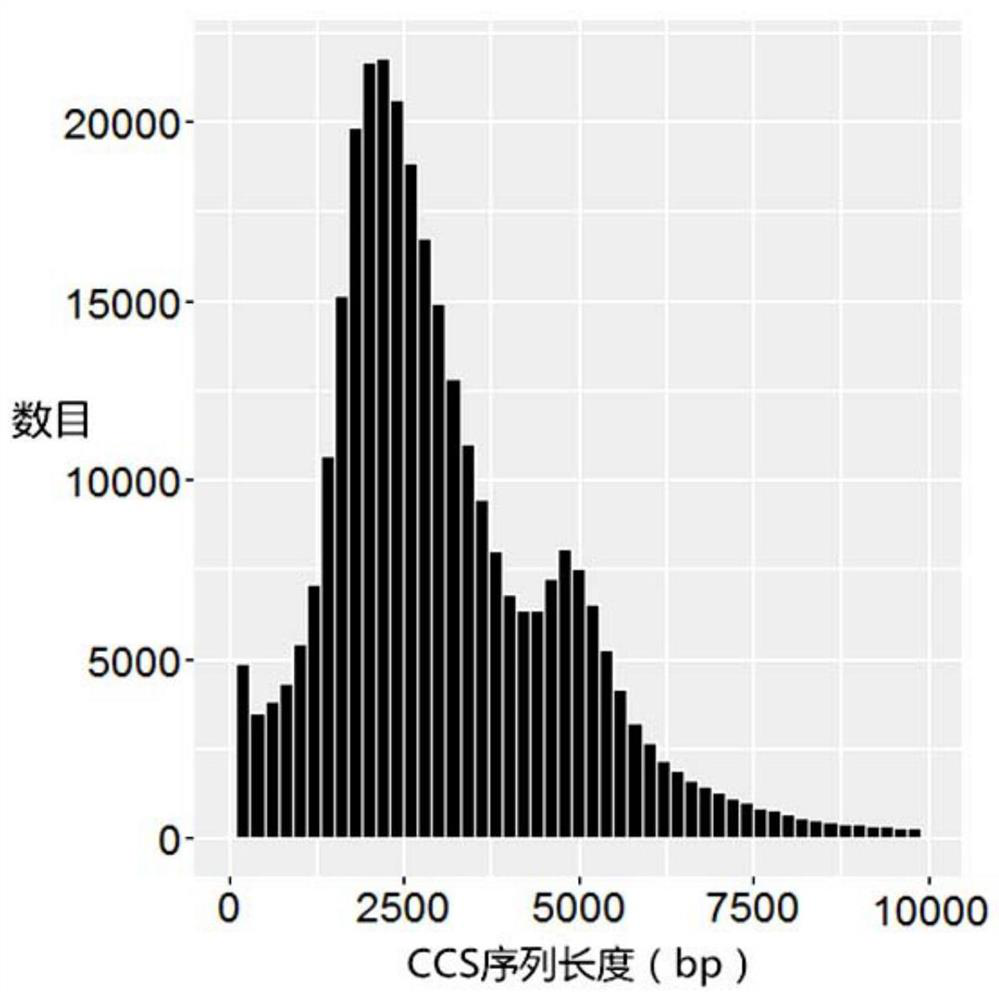
[0091] 2) Use the RS_ReadsOfInsert.xml protocol in SMRT Analysis2.3 to perform CCS correction (circular consensus sequencing) on the bax.h5 file (including polymerase read) of the PacBio offline data to obtain the fastq file. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com