Primer for performing double-PCR early and rapid detection on streptococcus agalactiae and streptococcus iniae as well as application thereof
A technology of Streptococcus iniae and Streptococcus nisella, applied in the field of microbial detection, can solve the problems of high cost, missed detection or false detection, false positive, etc., and achieve the effect of long solution cycle, low detection cost and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 The establishment of a double PCR method for rapid detection of Streptococcus agalactiae and Streptococcus dolphin
[0046] (1) Template preparation: select Streptococcus agalactiae, Streptococcus inus, Aeromonas hydrophila, Edwards tarda, Aeromonas victorini, Vibrio vulnificus, and mild from the strains stored at -80°C Aeromonas, Vibrio alginolyticus, and Escherichia coli were inoculated on the brain heart infusion agar medium under aseptic conditions, and cultured at a constant temperature at 28°C. After 24 hours, the cultured bacteria were collected and cultured respectively. Refer to Gram-positive bacteria DNA extraction method extracts the genomic DNA of various bacteria, and uses the DNA extraction kit (Dalian Bao Biological Engineering Co., Ltd.) to extract the genomic DNA and use it as a reaction template;
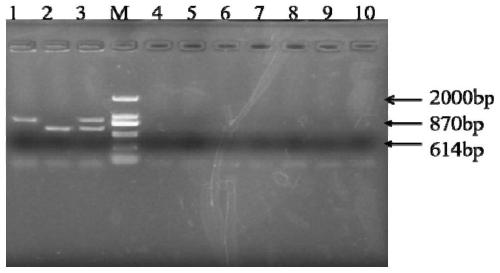
[0047] (2) Primers design and synthesis: According to the 16S rDNA partial sequence of Streptococcus agalactiae in GenBank, primers P-1 and P-2 are des...
Embodiment 2
[0056] Example 2 The detection of DNA sensitivity of Streptococcus agalactiae and Streptococcus dolphin by double PCR includes the following steps:
[0057] (1) Take 2-3 mL of activated Streptococcus agalactiae and Streptococcus dolphinus respectively, extract DNA with a commercial DNA extraction kit, and measure its concentration with a spectrophotometer;
[0058] (2) Dilute the genomic DNA of Streptococcus agalactiae to 9.84×10 -2 , 9.84×10 -3 , 9.84×10 -4 , 9.84×10 -5 , 9.84×10 -6 ng / μL, respectively dilute the genomic DNA of Streptococcus inus to 9.30×10 -2 , 9.30×10 -3 , 9.30×10 -4 , 9.30×10 -5 , 9.30×10 -6 ng / μL.
[0059] (3) such as figure 2 As shown, the DNA sensitivity of Streptococcus agalactiae and Streptococcus dolphin are 9.84×10 -5 ng / μL and 9.30×10 -5 ng / μL.
Embodiment 3
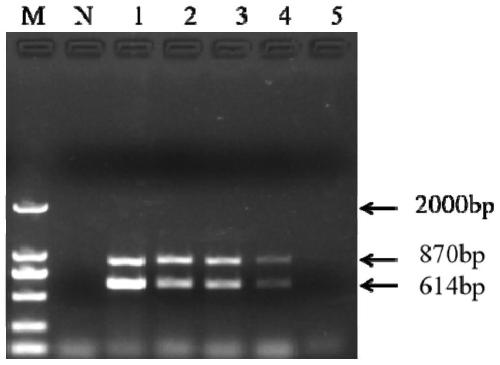
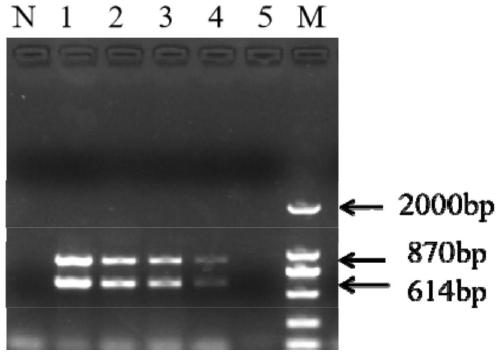
[0060] Example 3 Double PCR detection of single bacteria sensitivity of Streptococcus agalactiae and Streptococcus dolphin includes the following steps:
[0061] (1) Take the activated Streptococcus agalactiae and Streptococcus dolphin bacteria respectively, wash twice with 0.9% saline, dilute by ten times gradient, and count on the plate;
[0062] (2) Dilute the concentration of Streptococcus agalactiae to 2.76×10 respectively 6 , 2.76×10 5 , 2.76×10 4 , 2.76×10 3 , 2.76×10 2 , 27.6cfu / mL, dilute the concentration of Streptococcus dolphin to 2.51×10 6 , 2.51×10 5 , 2.51×10 4 , 2.51×10 3 , 2.51×10 2 , 25.1cfu / mL.
[0063] (3) Using the system and procedure of Example 1, boil the bacterial liquid prepared above for 10 minutes, and take the supernatant as a template for the sample to be tested;
[0064] (4) such as image 3 As shown, the sensitivity of pure bacteria of Streptococcus agalactiae and Streptococcus dolphin are 2.76×10 2 cfu / mL and 2.51×10 2 cfu / mL.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com