Protein-protein interaction prediction method using multivariate mutual information and residue binding energy
A prediction method and mutual information technology, applied in proteomics, informatics, bioinformatics, etc., can solve problems such as differences in prediction results, and achieve the effect of excellent accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
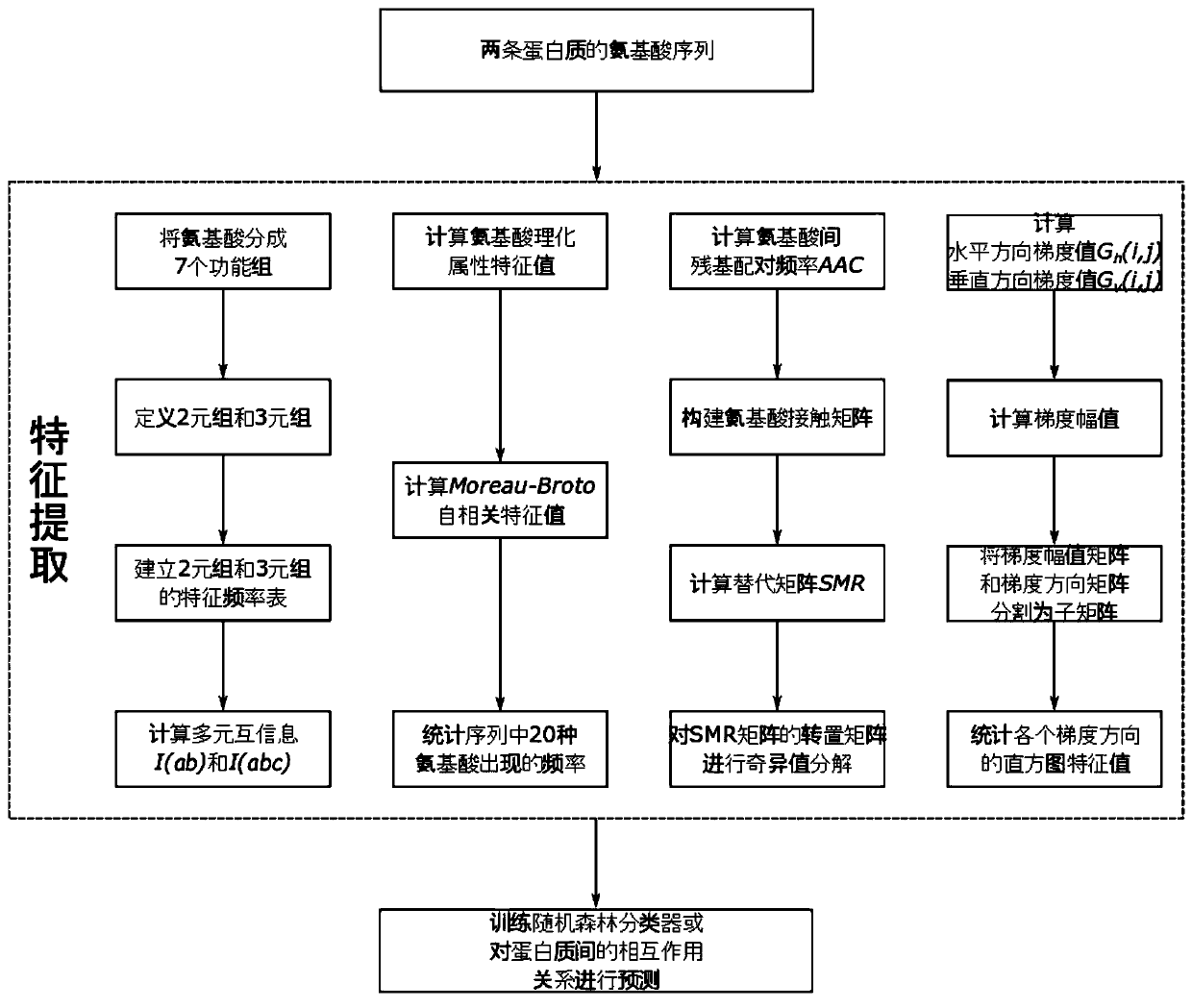
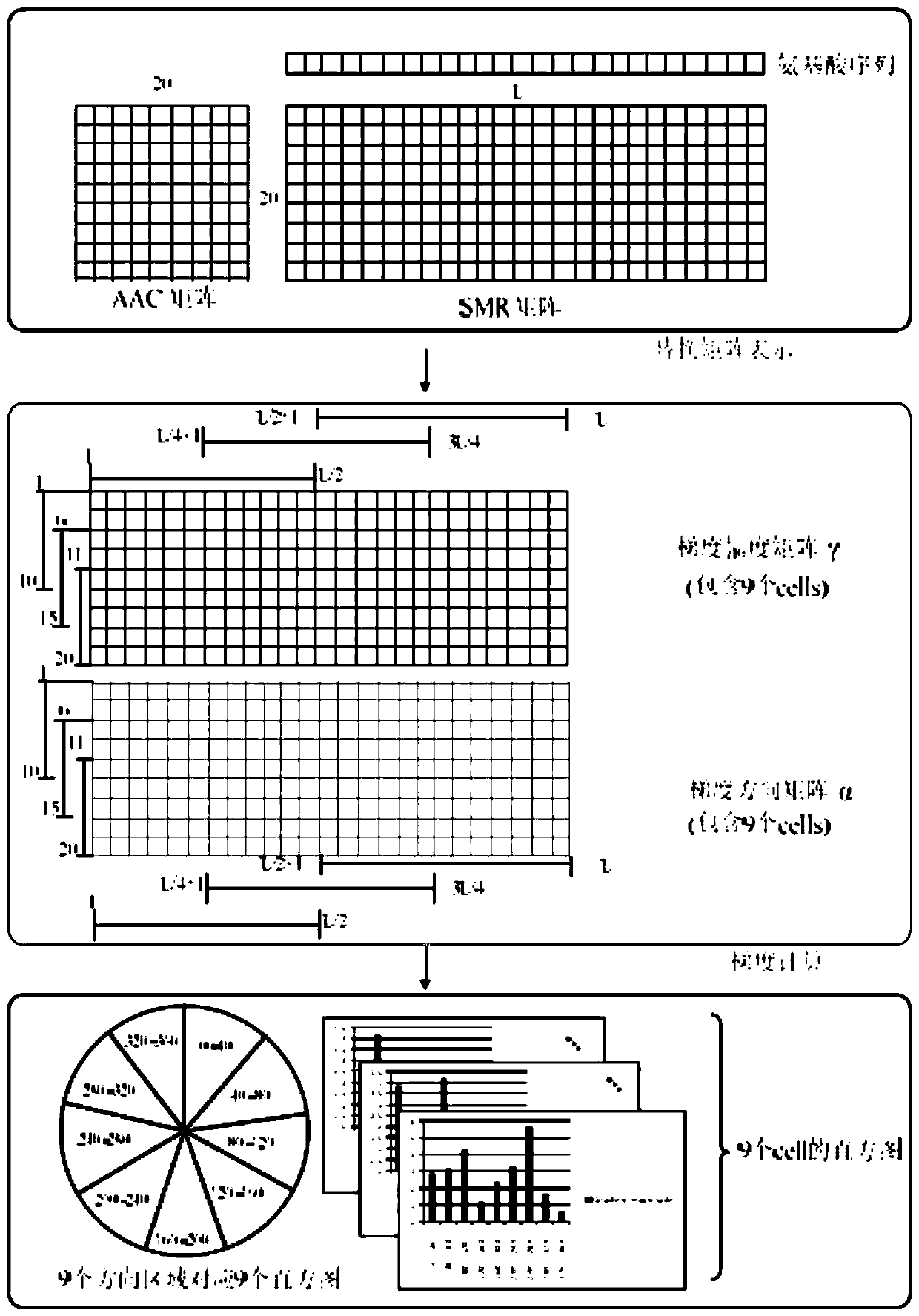
[0048]The purpose of the present invention is to provide a method for accurately and efficiently predicting the interaction between proteins. The feature extraction function used in the method can improve the role of useful information in the amino acid sequence in predicting operations, while effectively reducing the impact of useless noise information.
[0049] The present invention is characterized in that it contains the following steps successively:
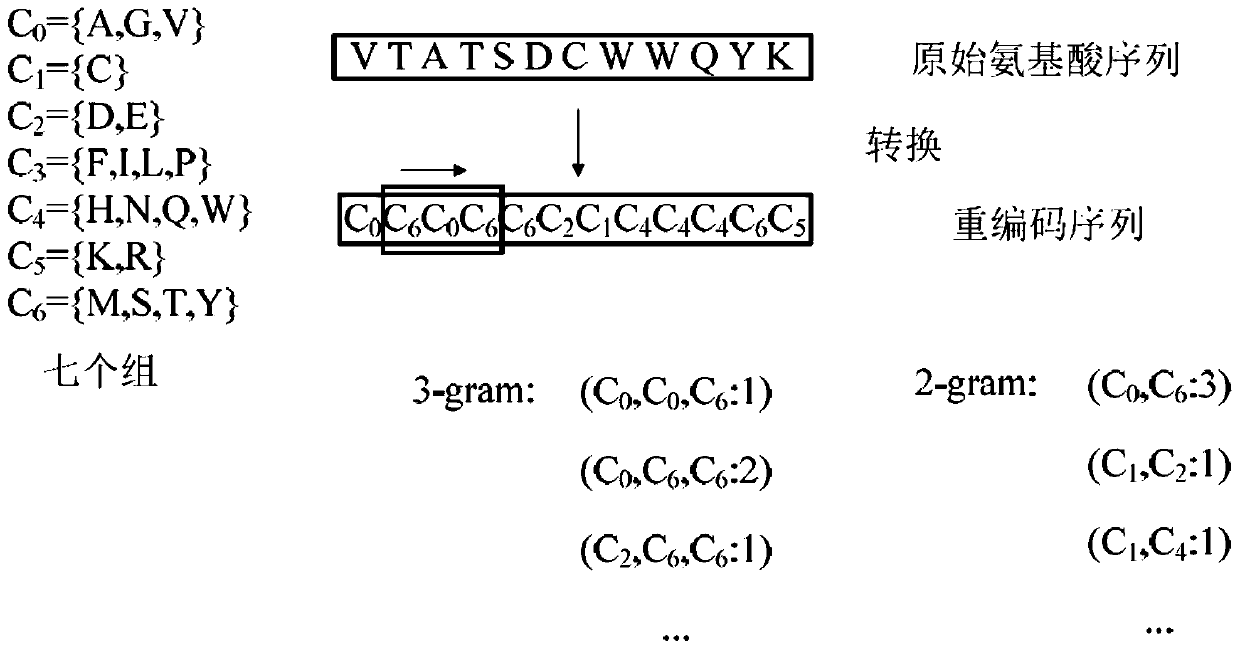
[0050] Step (1): Amino acid category grouping. The 20 standard amino acids were divided into 7 functional groups according to their dipolarity and volume. These seven functional groups are denoted as C 0 , C 1 , C 2 ,...,C 6 . The original amino acid sequence is converted into a group category sequence according to the functional group category of each amino acid.
[0051] Step (2): Define different types of 3-tuple and 2-tuple feature representations. The feature of the 3-tuple is expressed as "C 0 C 0 C 0 ","C ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com