Reagent kit for detecting diabetes
A diabetes and kit technology, applied in the field of detection, can solve the problems of less research on pyruvate dehydrogenase phosphatase and unclear functional localization, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Sample Selection and DNA Extraction
[0022] 1.1 Object
[0023] From January 2016 to December 2017, 120 patients with type 2 diabetes treated in Yichun People's Hospital were collected as the experimental group; 120 normal people who underwent physical examination during the same period were selected as the control group. The subjects in the two groups were all Chinese Han nationality, male and female Half, and there was no statistically significant difference between ages (P>0.05). All subjects had no family history of other genetic diseases and tumor history. There was no genetic relationship among the subjects, and all subjects gave informed consent.
[0024] 1.2 Specimen source
[0025] 2 mL of peripheral blood was collected from the two groups of subjects, anticoagulated with sodium citrate and stored in a low-temperature refrigerator at -70 °C.
[0026] Genomic DNA is extracted from whole blood, and the extraction method and steps of genomic DNA are a...
Embodiment 2
[0033] Embodiment 2PCR amplification detection
[0034] 1.1 Design and synthesis of primers
[0035] Primers are specific primers and probes that can detect the SNP site through numerous designs and optimizations:
[0036] Upstream primer sequence: 5'-atacattttttagcctaa-3',
[0037] Downstream primer sequence: 5'-gatctccttatttaaacta-3',
[0038] Probe: 5'-gtaattgaaagattttctatttt-3'; the 3' end of the probe is blocked to avoid extension during the reaction, and the primers are synthesized by Shanghai Shenggong Company.
[0039] 1.2 HRM typing based on quantitative PCR
[0040] In the reaction system, the concentration ratio of upstream and downstream primers is 1:10, that is, in a 10 μL reaction system, the final concentration of upstream primers is 0.3 pmol,
[0041] The final concentration of downstream primers is 3 pmol, and the concentration of probe is 3 pmol. The amplification length is 300 bp, and the probe is specific and complementary to G. The total volume of PCR re...
Embodiment 3
[0055] Embodiment 3 kit preparation and actual detection
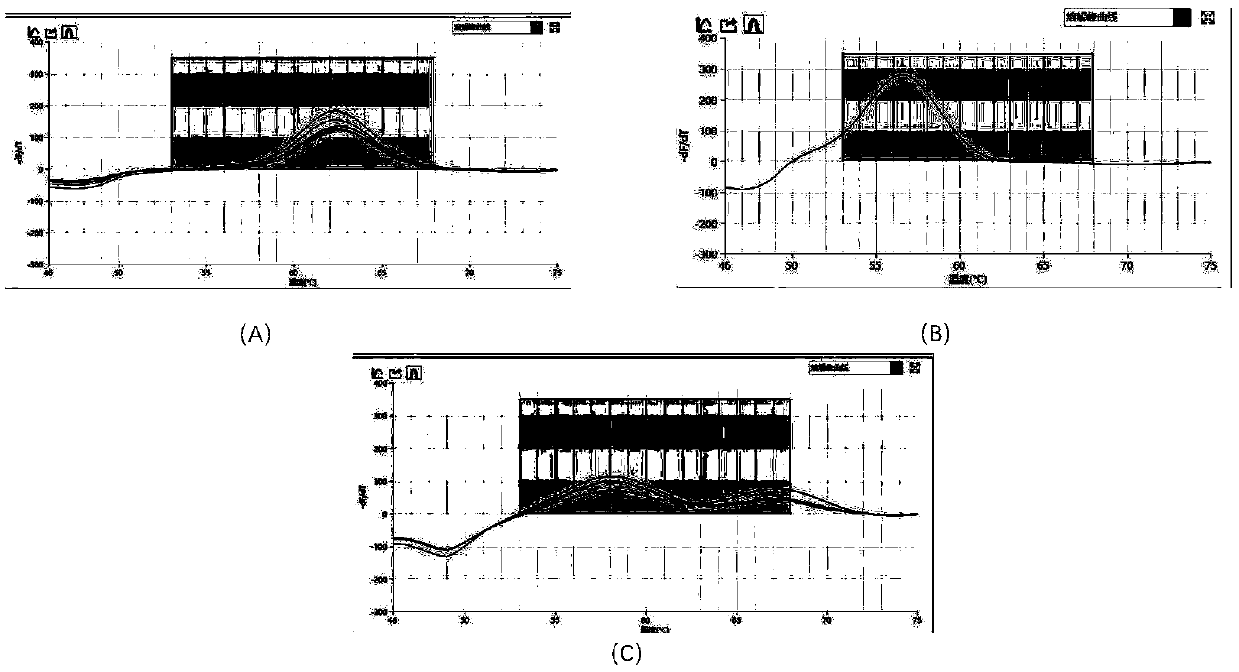
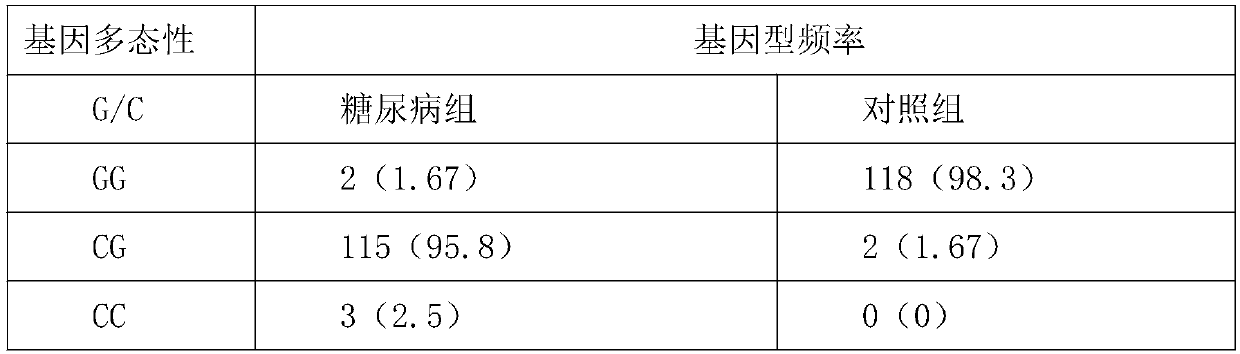
[0056] The probes and primers of the present invention are prepared into a kit according to the conventional storage concentrations of primers and probes in the art. Take 5 blood samples from diabetics and 6 blood samples from normal people, and carry out DNA extraction and PCR detection according to the same method as before. The curves of 11 samples are as follows: figure 1 As shown in the dissolution curve diagram of the test sample; there are 6 SNPs in the test samples with GG phenotype, 1 SNP with CC phenotype in total, and 4 cases with SNP in samples with CG phenotype, that is to say, CC phenotype plus CG A total of 5 cases were phenotyped, and the PCR test results also corresponded to the results of the samples being diabetic, indicating that the detection method has good accuracy.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com