Protein subnuclear localization method for feature extraction and fusion based on improved PSSM
A feature extraction and protein technology, applied in the fields of biology and information, can solve the problems of data set imbalance, low data set prediction accuracy, limitations, etc., to achieve the effect of enhancing complementarity and improving recognition rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
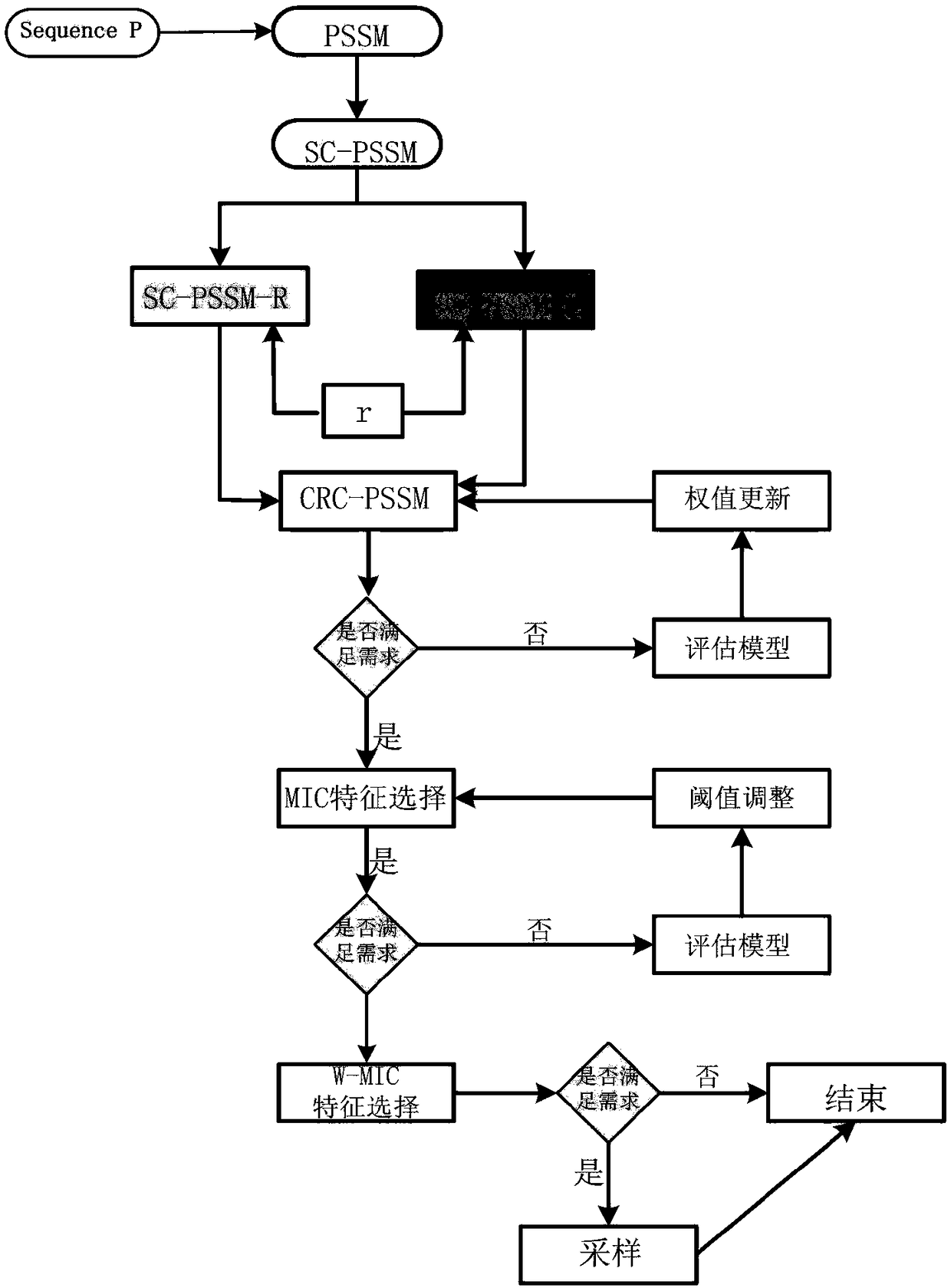
[0040] Such as Figure 1-2 Shown
[0041] A protein subnuclear localization method based on improved PSSM for feature extraction and fusion, including the following steps:
[0042] Step 1: Obtain a protein data set, determine whether the acquired data set is a single-label or multi-label problem (the present invention is mainly aimed at single-label problems), and convert the data set into a standard .fata format, and determine the category of all samples Make an annotation.
[0043] In step 1, a threshold is set for the acquired data set according to the length of each piece of data (generally the length is greater than 50) for data screening.
[0044] Step 2: Set the iteration parameter to 3, set the E-value value of each protein comparison search to 0.001, and calculate the PSSM matrix of each data. Each protein is represented by P, where P=[P1,P2,...,P20], Pj=[P1j,P2j,...PLj] (j=1, 2,...20), and L represents each The length of the protein.
[0045] Step 3: Convert the position s...
Embodiment 2
[0064] The present invention is based on the published apoptotic protein data set ZD98 for experimental verification. Among them, ZD98 was established by Zhou and Doctor in 2003. The data set contains four subcellular apoptotic protein sequences, namely cytoplasmic proteins (CY), plasma membrane-bound proteins (ME), mitochondrial proteins (MI) and other proteins (OTHER ). OA in Table 1 represents the overall correct recognition rate. The results in Table 1 strictly follow the feature extraction methods and fusion strategies mentioned above for feature fusion. In terms of feature selection, only traditional linear discriminant analysis algorithms are currently used for dimensionality reduction, and the results are better than traditional feature extraction methods. It can be seen from Table 1 that the algorithm in this paper is more effective than other algorithms in these evaluation objective indicators.
[0065] Table 1 Fusion results based on different fusion methods
[0066] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com