Method for detecting whether fusion mutation of target gene occurs, primer combination, kit and application thereof
A fusion mutation and primer combination technology, applied in the fields of biochemical equipment and methods, combinatorial chemistry, recombinant DNA technology, etc., can solve the problems of low capture efficiency, false negatives, difficult and unknown breakpoint fusion gene detection, etc., and achieve the enrichment effect. Good results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] This embodiment designs primer combinations that can be used to detect ALK fusion gene, NTRK1 fusion gene, RET fusion gene, ROS1 fusion gene, FGFR3 fusion gene and NTRK3 fusion gene, and its design principles are as follows:
[0062] (1) Select a target gene that may be fused with other genes, and determine the DNA region on the target gene that may be broken and fused;
[0063] (2) predicting the relative positional relationship between the other genes and the target gene after the cleavage fusion occurs, and designing a primer set according to the relative positional relationship:
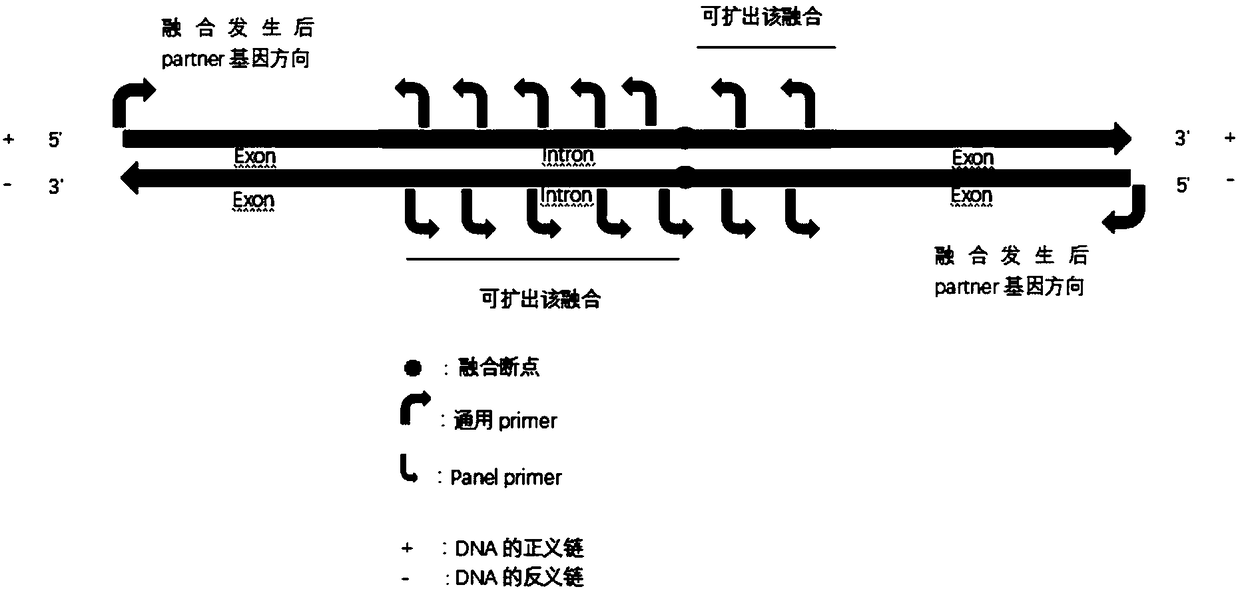
[0064] a. If after fusion, the other genes are located at the 5' end of the target gene, then sequentially design multiple 3' ends and the sense strand along the direction from the 3' end to the 5' end of the sense strand of the DNA region The primers with reverse complementary pairing of the strands constitute primer set 1, wherein, in each primer, the region that is reversely complementa...
Embodiment 2
[0073] The primer combination described in the above table 1 was used to establish the amplification library for sequencing. The specific library construction method is as follows, Figure 2 to Figure 13 The primer combination shown was outsourced and named as the library construction primer set. Figure 14 The general primers / upstream primers shown were synthesized outsourced, and other reagents used for library construction were purchased from Qiagen:
[0074] 1. Fragmentation, end repair and "A" ligation
[0075] 1.1. According to the sample type and sample quality, take 20-80ng of DNA for subsequent library construction operations. Among them, it is recommended to input 20-40ng for cfDNA samples and 40-80ng for FFPE samples. On ice, prepare fragmentation, end repair, and "A" ligation mixes according to Table 2. Centrifuge briefly, then pipette 7-8 times to mix and centrifuge briefly again.
[0076] Table 2 Fragmentation, end repair and "A" ligation reaction mix
[0077...
experiment example 1
[0130] In this experimental example, the performance of the optimized primer combination and library construction method finally optimized in Example 1 of the present invention was firstly verified by using the internationally recognized Horizon standard. Build a library and use Illumina's Nextseq550AR sequencer to sequence the constructed library and analyze the off-machine data, and count whether the detected mutation results are consistent with the results marked by the manufacturer. Secondly, in addition to standard products, we also verify the negative and positive clinical samples collected on the market and verified by other testing platforms, and count the consistency of the results. In addition, considering that there are few amplicon-based sequencing kits in the prior art to detect fusion mutations at the DNA level, this experimental example chooses a commercialized Qiagen non-specific gene mutation detection kit for fusion mutations (Lung Cancer Panel), as a control...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com