Method for quantitative PCR detection of maspin gene methylation state and application thereof
A methylation and non-methylation technology, which is applied in the fields of biotechnology and medicine, can solve problems such as difficult to detect, inability to detect early in situ breast cancer, and inability to meet tumor molecular typing, so as to avoid accidental errors, Optimizing the PCR system and procedures to ensure the effect of accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
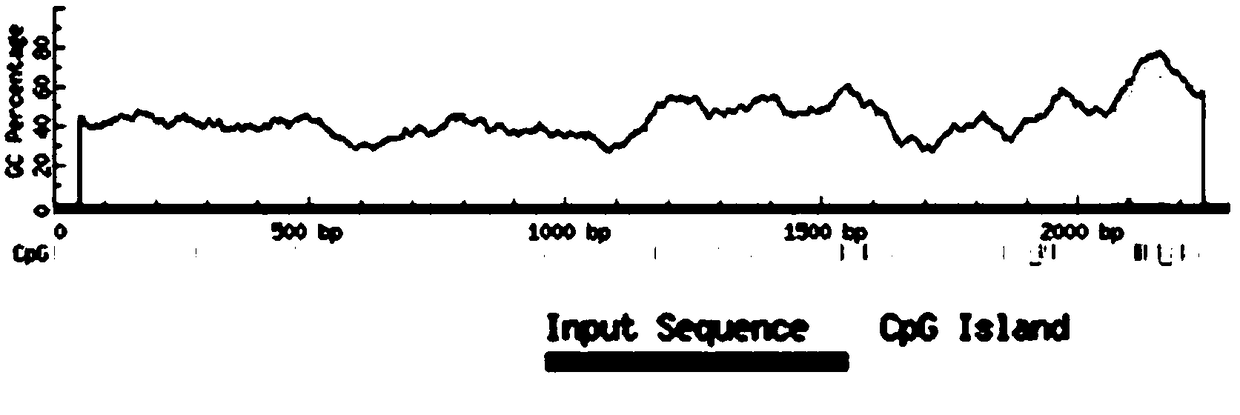
[0045] Example 1: Determine the specific site and sequence information of the CpG island in the upstream regulatory region of the maspin gene coding sequence
[0046] The breast cancer detection method of the present invention is based on the principle of methylation-specific PCR. After the single-stranded DNA is modified by bisulfite, all unmethylated cytosines are deaminated into uracils, and the methyl groups in the CpG sites are The modified cytosine remains unchanged, and the formazan can be amplified by PCR
[0047] Differentiate between methylated and unmethylated DNA sequences. Therefore, it is necessary to first determine the specific site and sequence information of the CpG island in the upstream regulatory region of the maspin gene coding sequence.
[0048] Use the gene sorter tool at http: / / genome.ucsc.edu / index.html, select the species as human, select the latest database, search for the maspin gene, select the upstream 2000bp and 5'UTR of the promoter, and obtai...
Embodiment 2
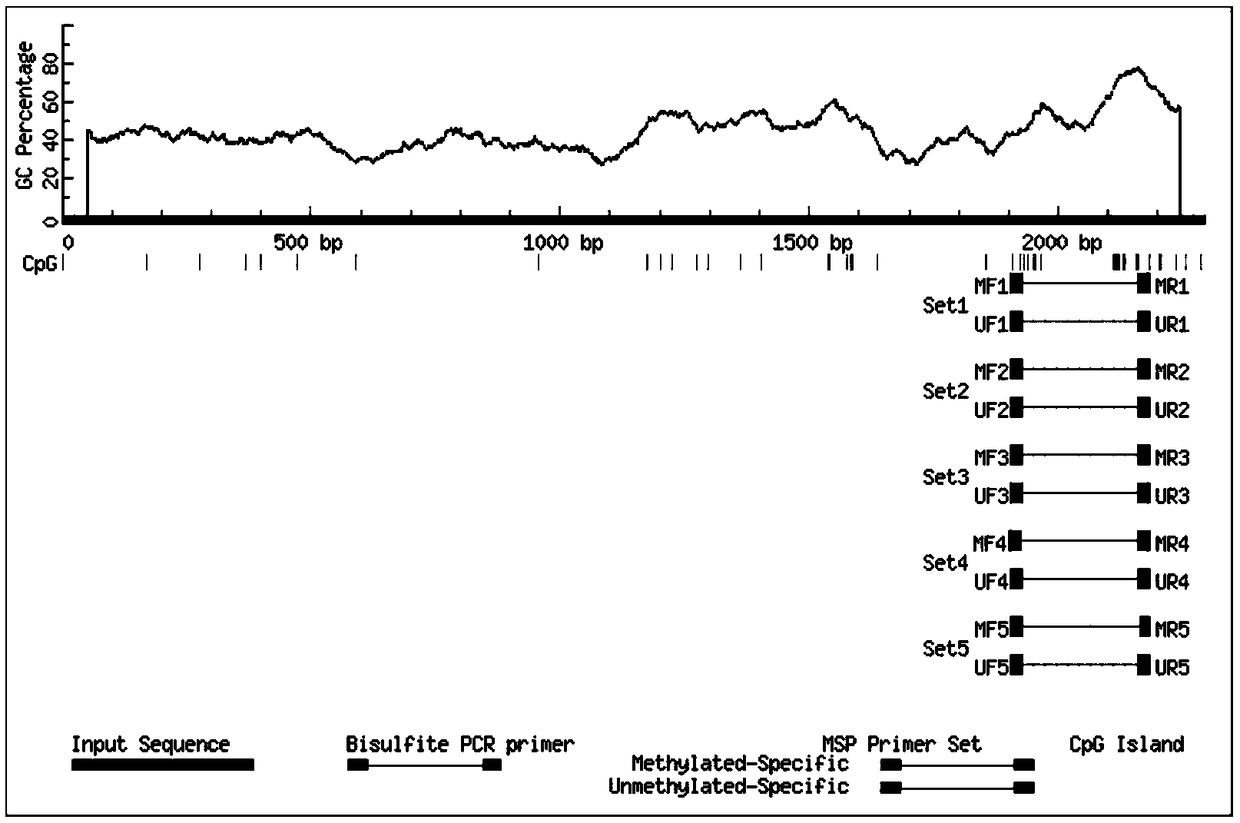
[0049] Example 2: Design and synthesis of primers for specifically amplifying the CpG island of the upstream regulatory region of the maspin gene coding sequence
[0050] According to the CpG island information of the upstream regulatory region of the maspin gene coding sequence in Example 1, two sets of primers for methylated and unmethylated sequences were designed, and these 10 pairs of primers can be used to specifically amplify the upstream regulation of the maspin coding sequence The region contains methylated and unmethylated sequences of different lengths of CpG islands, and subsequent methylation quantitative analysis is performed to specifically detect the methylation status of the corresponding CPG site. According to the sequence shown in SEQ ID NO.1, go through the http: / / www.urogene.org / cgi-bin / methprimer / methprimer.cgi website, select the Pick MSP primers option, and check Use CpG island prediction for primer selection? Then click submit to get the primers used t...
Embodiment 3
[0051] Embodiment 3: establish the method for detecting the methylation state of maspin gene based on quantitative PCR
[0052] The detection method of the present invention is based on the principle of methylation-specific PCR, and primers for methylated and unmethylated sequences are designed respectively, and methylated and unmethylated DNA sequences can be distinguished through PCR amplification . The method comprises the steps of:
[0053] 1. Extract genomic DNA from tissue samples and perform quality inspection on the extracted DNA samples
[0054] According to the diagnostic needs of whether the patient has breast cancer metastasis and malignancy, the tissue sample can be a small amount of breast cancer clinical tissue samples taken out minimally invasively, or it can be a breast cancer tissue sample from the metastatic site of breast cancer; for normal human breast cancer For the needs of early screening such as morbidity, tissue samples can be a small amount of brea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com