Protein-DNA binding residue prediction method based on sampling and integrated learning
A prediction method and technology for binding residues, applied in proteomics, genomics, informatics, etc., can solve the problems of low prediction accuracy and information loss of the final model, enrich feature sources, prevent overfitting, reduce The effect of information loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
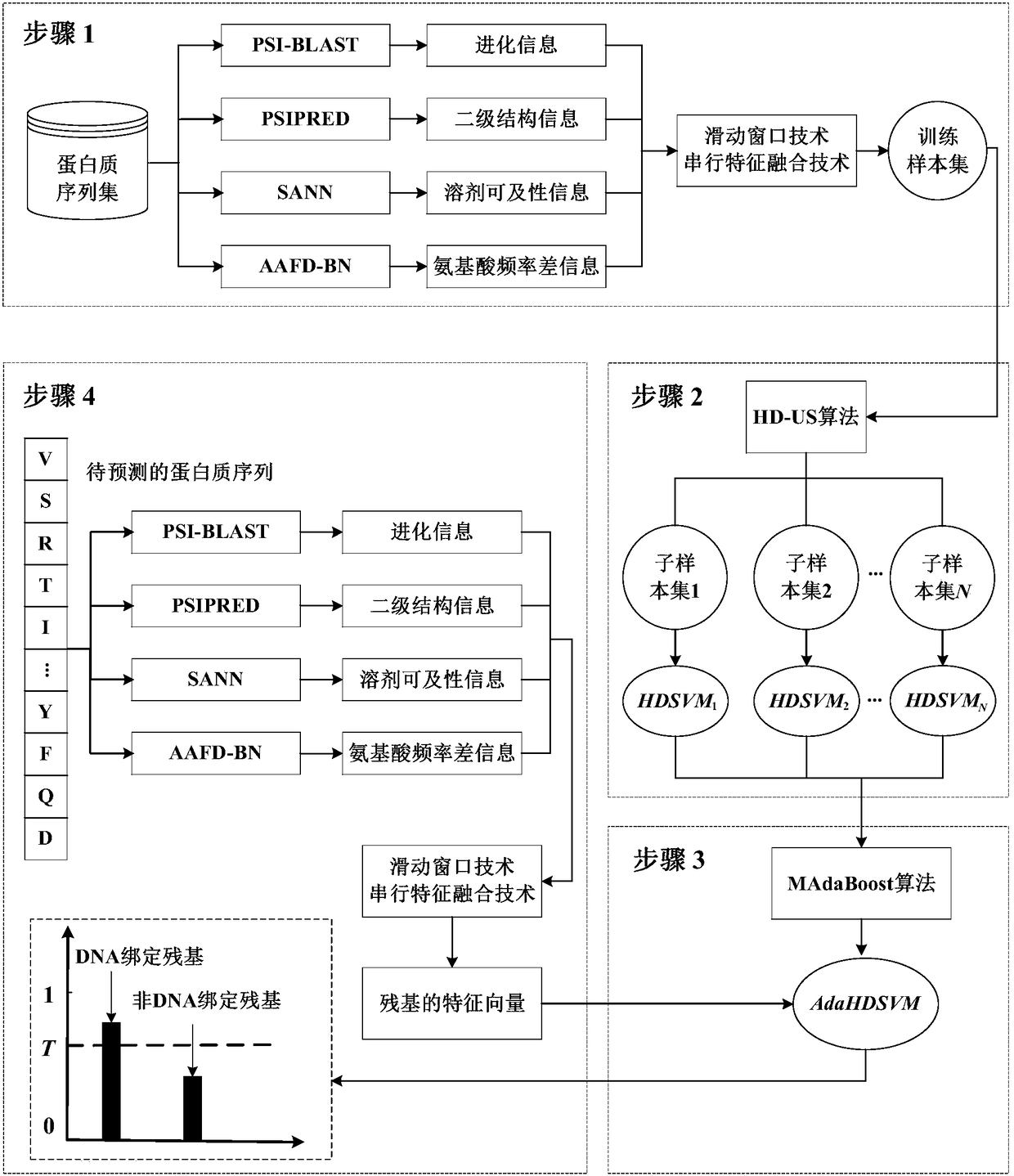
[0015] The present invention will be further described below in conjunction with the accompanying drawings.
[0016]The accompanying drawing shows a schematic structural diagram of the prediction method system of the present invention. As shown in the accompanying drawings, according to an embodiment of the present invention, a method for predicting protein-DNA binding residues based on sampling and integrated learning includes the following steps: First, given a protein sequence set, use PSI-BLAST, PSIPRED, SANN and AAFD-BN algorithms extract the evolution information, predicted secondary structure information, predicted solvent accessibility information and amino acid frequency difference information of each protein sequence respectively; on this basis, combined with sliding window technology and serial The feature fusion technology represents the amino acid residues in the sequence in the form of feature vectors, and constructs a training sample set in units of residues. S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com