Method and device for filling holes based on third-generation sequencing sequences
A sequence and sequencing technology, applied in the field of biological information, can solve the problems of slow sequence alignment and high resource consumption, and achieve the effect of improving speed and saving consumption.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
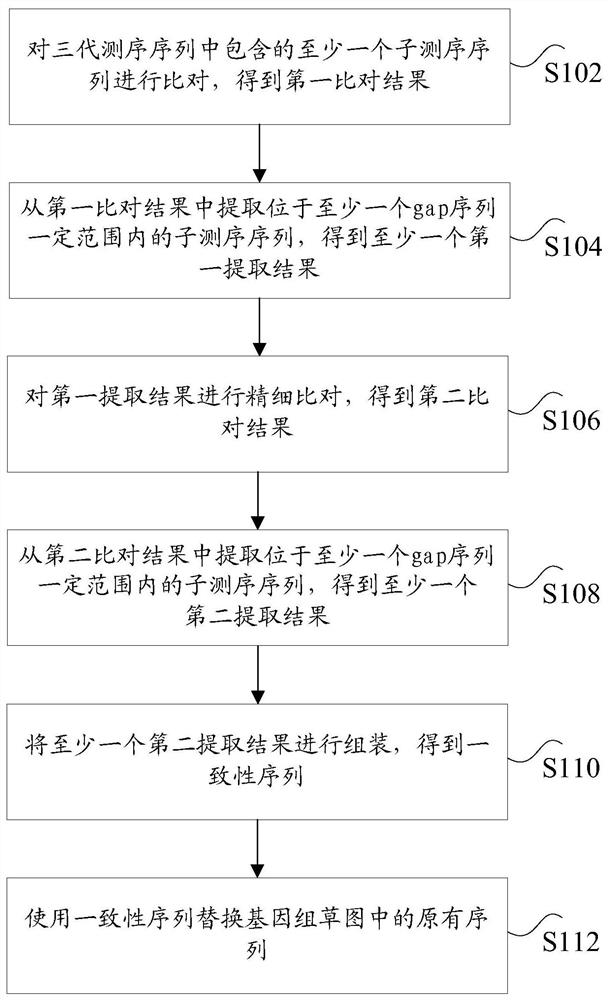
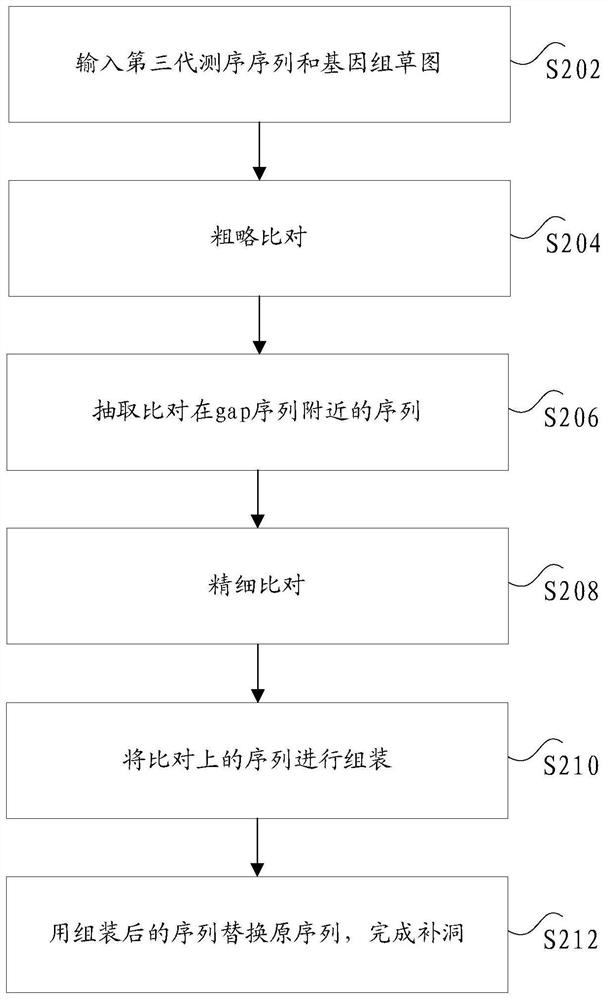
[0034] In the related art, only unique comparison software is adopted. The current alignment software for long sequences, fast and precise
[0039] For example, Minimap2 software can be used to quickly align third-generation sequencing reads.
[0045] For example, the second alignment software can be a bedtools tool, using the bedtools tool to compare genomes with grass
[0046] The original reads are divided into two categories: the first category is the three-generation sequencing reads that are aligned to the gap sequence within a certain range,
[0055] Optionally, step S112 includes: using a preset number of consistent sequences to replace the corresponding gap sequences.
[0059] Specifically, looking for gap sequences in the draft genome, and defining consecutive 25 or more Ns as gap sequences.
[0068]
[0070]
[0071] Through the real data test, the hole-filling effect of the application is slightly better than PBjelly, and the comparison speed can be improved by 8-10 time...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com