Extracting kit for formalin soaked tissue DNA and extraction method
A formalin and kit technology, applied in the field of DNA extraction, can solve the problems of difficulty in obtaining, long extraction time, and inability to break cells well, and achieve high recovery rate, good DNA quality, and improved DNA recovery rate. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Formalin Soaked Tissue DNA Extraction Kit
[0053] 1. Preparation of SDS buffer: Take 1g of SDS, add 50mM Tris-HCl (pH8.1) and mix well, and dilute to 100ml.
[0054] 2. Preparation of binding buffer: Weigh 31.84g GuHCl, 4.38g NaCl, 0.42g SDS, 70g Triton X-100, add 80ml ultrapure water, mix well, add 0.5ml EDTA, 5ml Tris-HCl, and dilute to 100ml.
[0055] 3. Preparation of 1.28mol / LEDTA: Weigh 18.61g EDTA into a 50mL beaker, and add 40mL ultrapure water. Place the beaker on a magnetic stirrer, add NaOH solid while stirring until the solution becomes clear, adjust the pH to 8.0, and dilute to 50 mL.
[0056] 4. 1mol / LTris-HCl (pH 8.1) solution preparation: Weigh 12.11g Tris into a 100mL beaker, and add 80mL ultrapure water to completely dissolve it, measure the pH with a pH meter, and adjust the pH with concentrated hydrochloric acid. Transfer the solution to a 100mL volumetric flask and add purified water to make the volume up to 100mL.
[0057] 5. Preparation of the first wash...
Embodiment 2
[0061] Pretreatment
[0062] 1. Select the lung tissue of a cancer patient soaked in formalin, take the tissue with an 18-gauge puncture needle, use a scalpel blade to cut the tissue into granules with a diameter of 1mm, and divide them into 5 samples with a weight of 4.2mg. . Open the screw cap of microTUBE, add 100μl SDS Buffer, add the minced tissue to the Buffer, tighten the cap, and number 1-5 respectively.
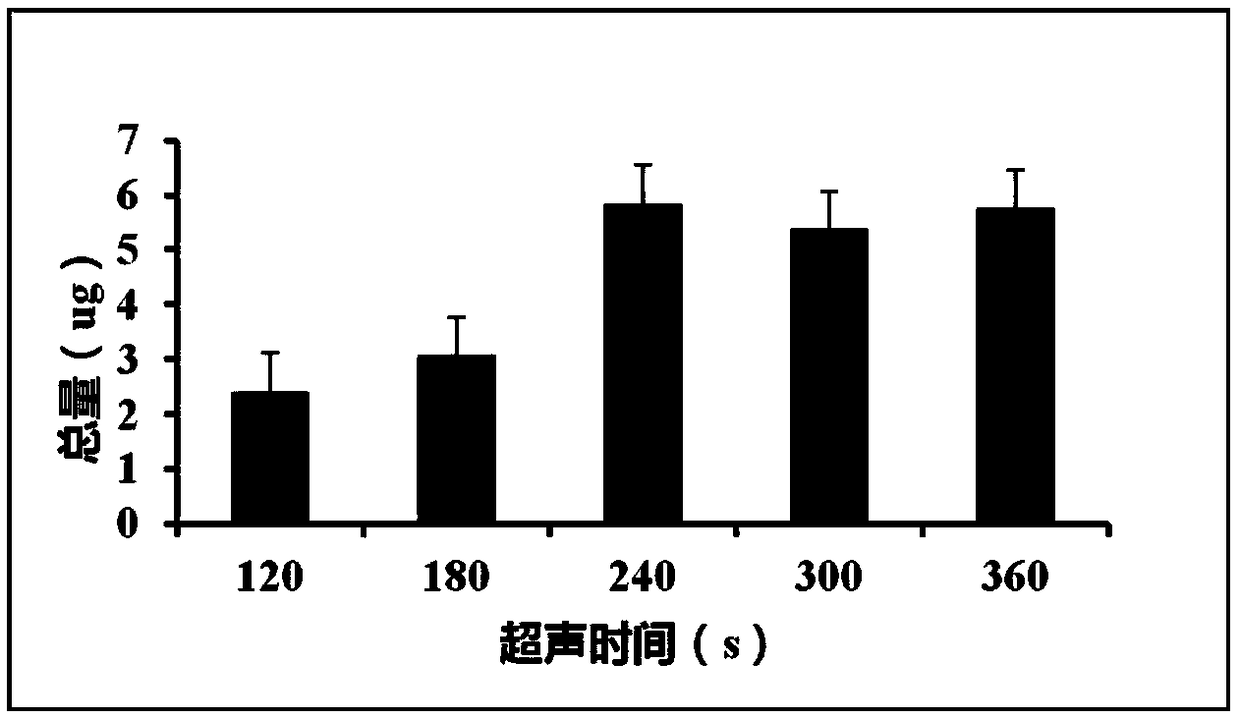
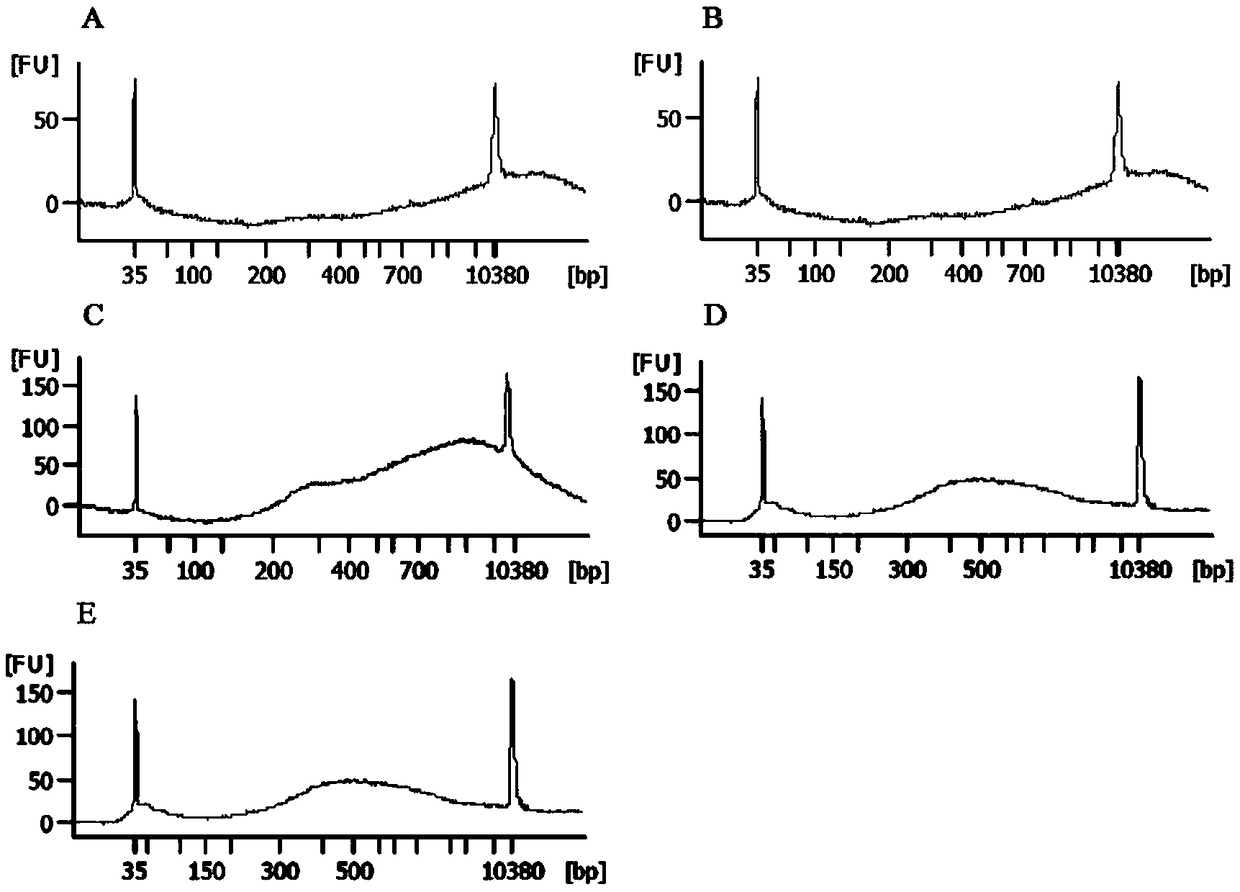
[0063] 2. Turn on the ultrasonic disruptor and set up the program. Under the condition that other parameters are the same, the fragmentation time of samples 1-5 increases in order, respectively, 120s, 180s, 240s, 300s, 360s. Samples No. 1 and No. 2 can be seen to have a small amount of tissue mass, which is not completely homogenized, and the other three groups are completely homogenized. See Table 1 for ultrasonic breaking parameters.
[0064] Table 1 Ultrasonic breaking parameters
[0065]
[0066] 3. Open the lid and pipette the sample from the microTUBE into a 1.5ml E...
Embodiment 3
[0088] 1. Select a lung tissue of a cancer patient soaked in formalin, take the tissue with an 18-gauge puncture needle, use a scalpel to cut the tissue into granules with a diameter of 1 mm, and divide them into 5 samples with a weight of 3.9 mg. . Open the screw cap of microTUBE, add 100μl SDS Buffer, add the minced tissue to the Buffer, tighten the cap, and number 1-5 respectively.
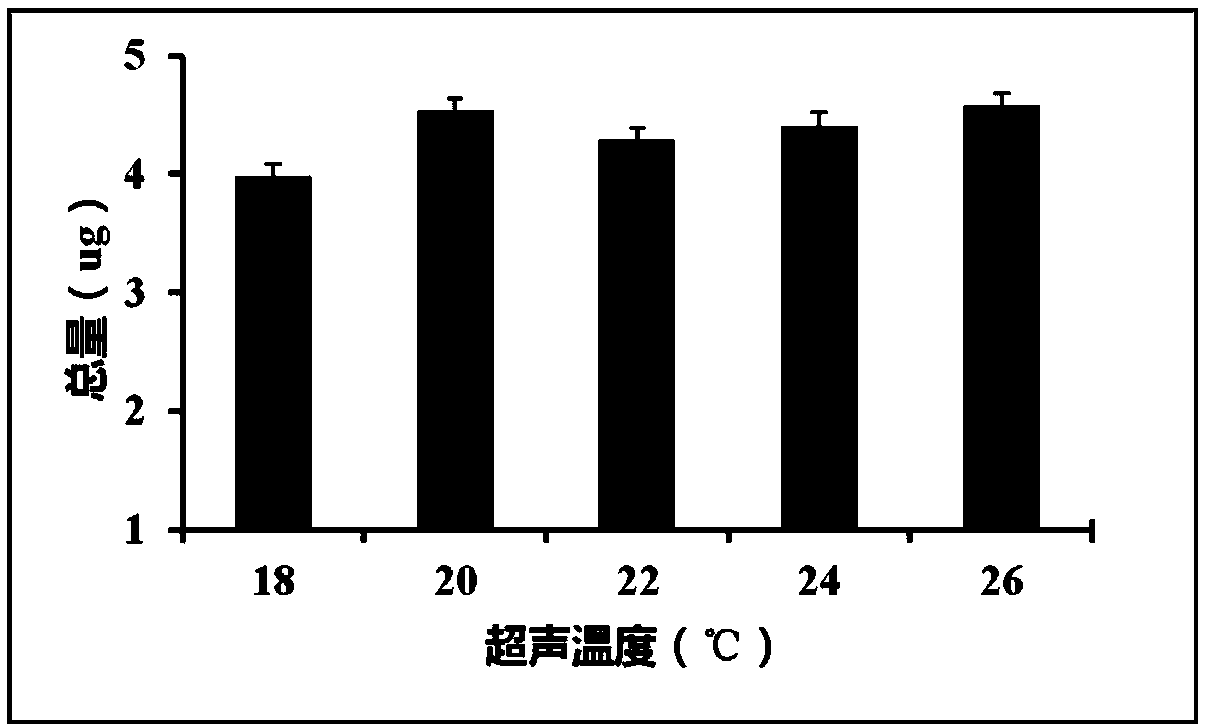
[0089] 2. Turn on the ultrasonic disruptor and set the program to disrupt the tissue. Under the condition that other parameters are the same, the disruption temperature of samples 1-5 increases sequentially, respectively 18℃, 20℃, 22℃, 24℃, 26℃. It can be seen that the 5 groups of samples are completely homogenized. See Table 3 for ultrasonic breaking parameters.
[0090] Table 3 Ultrasonic breaking parameters
[0091]
[0092] 3. Open the lid and pipette the sample from the microTUBE into a 1.5ml EP tube.
[0093] 4. Add 20μl proteinase K to the sample and mix well.
[0094] 5. Incubate at 56°C for ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com