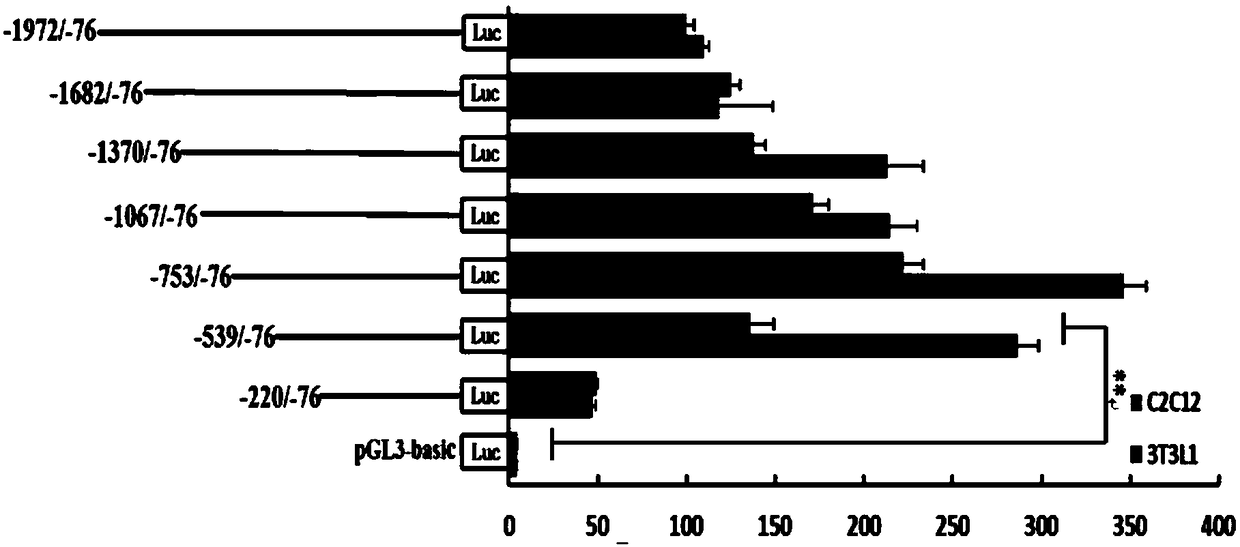
Method for identifying active region of ATP5B (adenosine triphosphate 5B) gene promoter
A technology of active regions and promoters, applied in biochemical equipment and methods, recombinant DNA technology, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] 1. Analysis of luciferase activity of ATP5B gene promoter
[0037] 1. Design of promoter primers
[0038] According to the 5′UTR sequence of the ATP5B gene in GeneBank, the transcription start site identified by 5′-RACE is +1, and gene-specific primers were designed by Clone Manage software:
[0039] The upstream primer is Forward Primer F1: 5'-CAGTCTTTCTCCTTTATTCCTCTGCTA-3'
[0040] The downstream primer is Reverse Primer R1: 5'-TCTGCTTATCACTCTGGGCG-3'.
[0041] Bovine genomic DNA was used as a template for PCR amplification, and the target fragment was 1897bp in total from the promoter sequence -1972 to -76. Add 1.2μL (100ng) DNA template, 0.4μL KOD-Plus-Ver 2.0 (1U / μL), 2μL 10×PCR Buffer, 2μL dNTP (2mmol / L), 0.8μL MgSO to a 200μL PCR tube 4 (25mol / L), 0.6μL ForwardPrimer F1 (10μM), 0.6μL Reverse Primer R1 (10μM), 12.4μL ddH 2O, a total of 20 μL, vortexed and centrifuged, placed in a PCR instrument for reaction, the reaction conditions are as follows: pre-denatura...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com