Specific primer and rapid detecting method for detecting lasiodiplodia theobromae
A detection method and cocoa ball technology, applied in the field of microbial detection, can solve the problems of long time, cumbersome procedures, delays, etc., and achieve the effects of good specificity, high sensitivity, economy and short time consumption.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1, specific primer design
[0032]According to the translation elongation factor gene (Transcription elongation factor, TEF) sequence of Botryobacteriaceae fungi including Lasiodiplodia theobromae and other species of fungi in GenBank, a variation interval that can be used to design L. theobromae-specific primers was found. The sequence is acgcatgtcgttttttaacccctctcga (i.e. SEQ ID NO: 1), the upstream primer ZYLTF1 for L.theobromae detection is designed using this interval sequence, and the downstream primer ZYLTR1 for L.theobromae detection is designed according to other base sequences of the translation elongation factor, which The nucleotide sequence is:
[0033] (SEQ ID NO: 2) ZYLTF1: 5'-acgcatgtcgttttttaa-3';
[0034] (SEQ ID NO:3) ZYLTR1: 5'-tacatgagtggtcagagcat-3'.
Embodiment 2
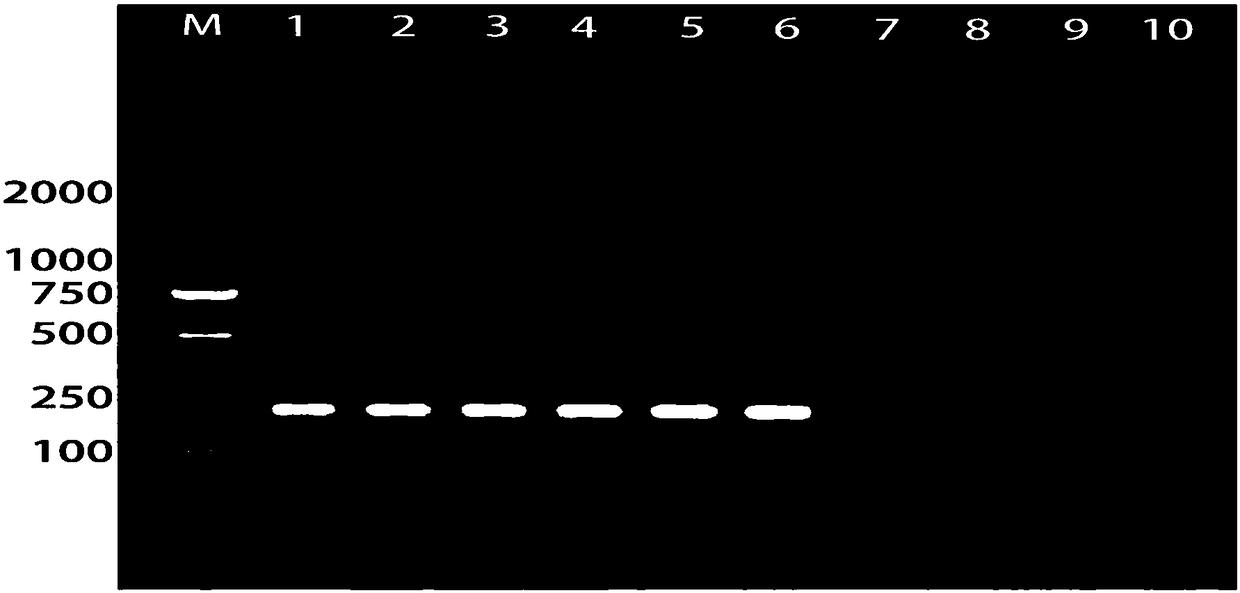
[0035] Example 2, a molecular detection method for rapid detection of phytopathogenic fungus Dispora coccidioides
[0036] 2.1 Test strains
[0037] Lasiodiplodia theobromae isolated from mango (strain LTMG1), L. theobromae isolated from mango (strain LTMG9), L. theobromae isolated from longan (strain LTLY13), L. theobromae isolated from longan (strain LTLY25), L. theobromae isolated from kiwi (strain MHT5), isolated from grape L. theobromae (strain LTPT2), isolated from cherry L. theobromae (strain LTYT42), isolated from bayberry L. theobromae (strain LTYM6), isolated from guava L. theobromae (strain LTFSL9), L. theobromae (strain LTXJ57) isolated from banana.
[0038] L.pseudotheobromae、L.crassispora、Neofusicoccum mangiferae、N.parvum、Botryosphaeria dothidea、Colletotrichum scovillei、Mortierella minutissima、Penicillium urticae、Acremonium furcatum、Truncatella angustata、Aspergillussydowii、Alternaria alternata、Phyllosticta capitalensis、Stemphyliumlycopersici、Pestalotiopsis mangi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com