Method for detecting genotype of polymorphic sites of corneal dystrophy gene and its kit
A technology of gene polymorphism and polymorphism loci, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of judgment of unfavorable results and high background, achieve accurate and reliable judgment of results, reduce pollution, easy results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
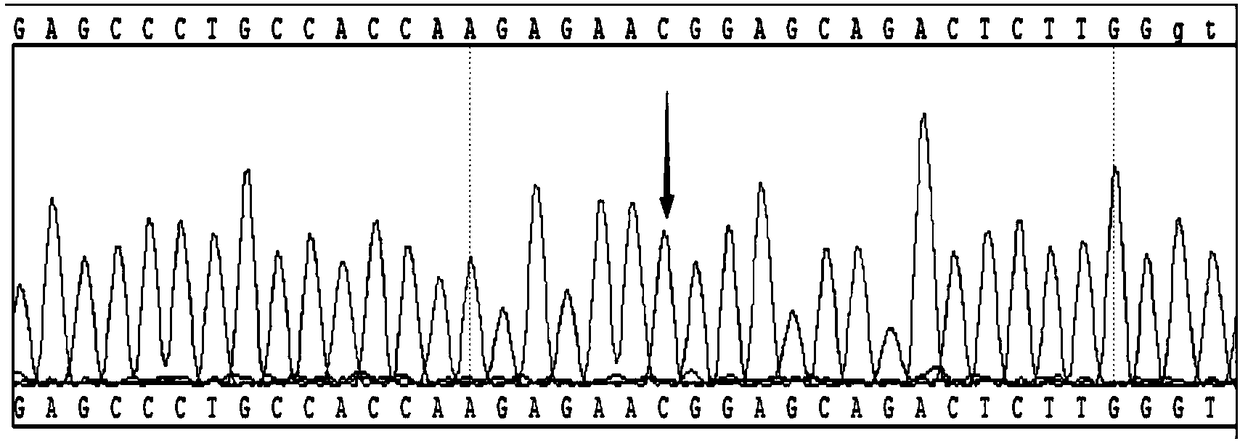
[0059] Example 1 Detection of the TGFBI gene R555W point mutation genotype related to corneal dystrophy
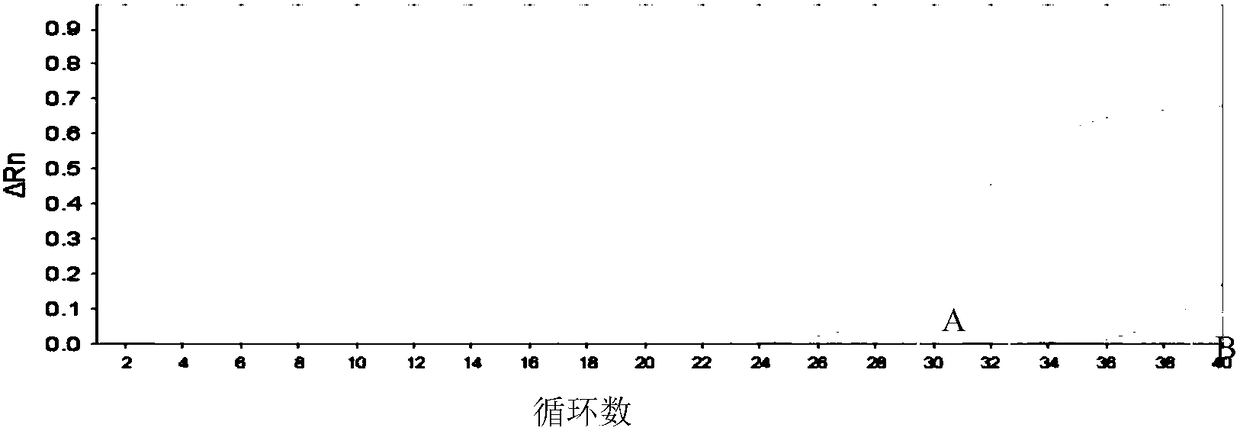
[0060] The present invention designs internal reference β-actin primers to evaluate whether the amplification of extracted samples is normal in real-time fluorescent quantitative PCR, making the results easier to judge.
[0061] The present invention uses enhanced ARMS primers to reduce the amplification efficiency of wild-type enhanced ARMS primers to mutant samples and the amplification efficiency of mutant enhanced ARMS primers to wild-type samples in real-time fluorescent quantitative PCR, making the results easier to judge .
[0062] 1) β-actin primer design
[0063] The upstream and downstream primers of β-actin gene were designed in the experiment. The relevant parameters are: Tm value 55.0℃-60.0℃, GC value 40.0%-60.0%, primer size 20±3bp. The primer sequences are as follows:
[0064] Upstream primer: β-actin-F AGTGGCTTCCCCAGTGTGACATG (SEQ ID NO: 1)
[0065] Dow...
Embodiment 2
[0085] Example 2 Detection of the genotype of the TGFBI gene R124H point mutation associated with corneal dystrophy
[0086] The present invention designs internal reference β-actin primers to evaluate whether the amplification of extracted samples is normal in real-time fluorescent quantitative PCR, making the results easier to judge.
[0087] The present invention uses enhanced ARMS primers to reduce the amplification efficiency of wild-type enhanced ARMS primers to mutant samples and the amplification efficiency of mutant enhanced ARMS primers to wild-type samples in real-time fluorescent quantitative PCR, making the results easier to judge .
[0088] 1) β-actin primer design
[0089] The upper and lower primers of β-actin gene were designed in the experiment. The related parameters are: Tm value 55.0℃-60.0℃, GC value 40.0%-60.0%, primer size 20±3bp. The primer sequences are as follows:
[0090] Upstream primer: β-actin-F AGTGGCTTCCCCAGTGTGACATG (SEQ ID NO: 1)
[0091] ...
Embodiment 3
[0112] Example 3 Detection of TGFBI gene R124L point mutation genotype related to corneal dystrophy
[0113] The present invention designs internal reference β-actin primers to evaluate whether the amplification of extracted samples is normal in real-time fluorescent quantitative PCR, making the results easier to judge.
[0114] The present invention uses enhanced ARMS primers to reduce the amplification efficiency of wild-type enhanced ARMS primers to mutant samples and the amplification efficiency of mutant enhanced ARMS primers to wild-type samples in real-time fluorescent quantitative PCR, making the results easier to judge .
[0115] 1) β-actin primer design
[0116] The upper and lower primers of β-actin gene were designed in the experiment. The related parameters are: Tm value 55.0℃-60.0℃, GC value 40.0%-60.0%, primer size 20±3bp. The primer sequences are as follows:
[0117] Upstream primer: β-actin-F AGTGGCTTCCCCAGTGTGACATG (SEQ ID NO: 1)
[0118] Downstream prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com