Application of LHAP1 protein and its coding gene in regulation of plant photosynthesis
A technology for photosynthesis and transgenic plants, which is applied in the fields of application, plant peptides, and plant products, and can solve the problems of in-depth research on interrelationships and influencing factors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Embodiment 1, the acquisition of LHAP1 gene
[0074] 1. Mutant acquisition and phenotype
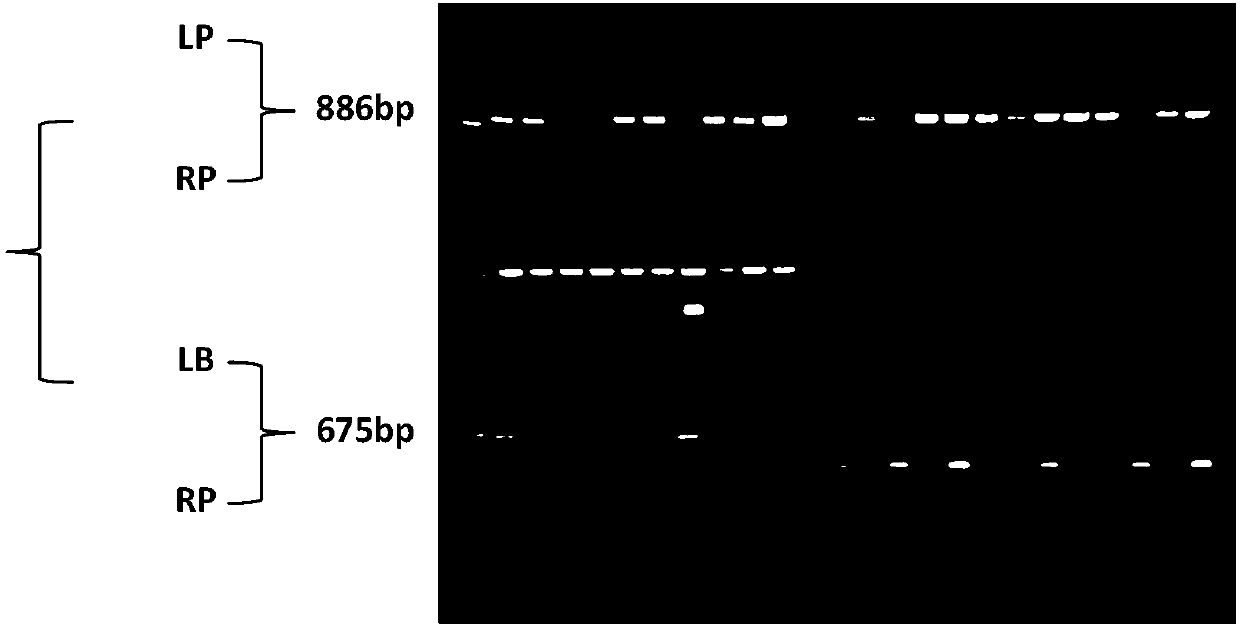
[0075] 1. Obtaining mutants
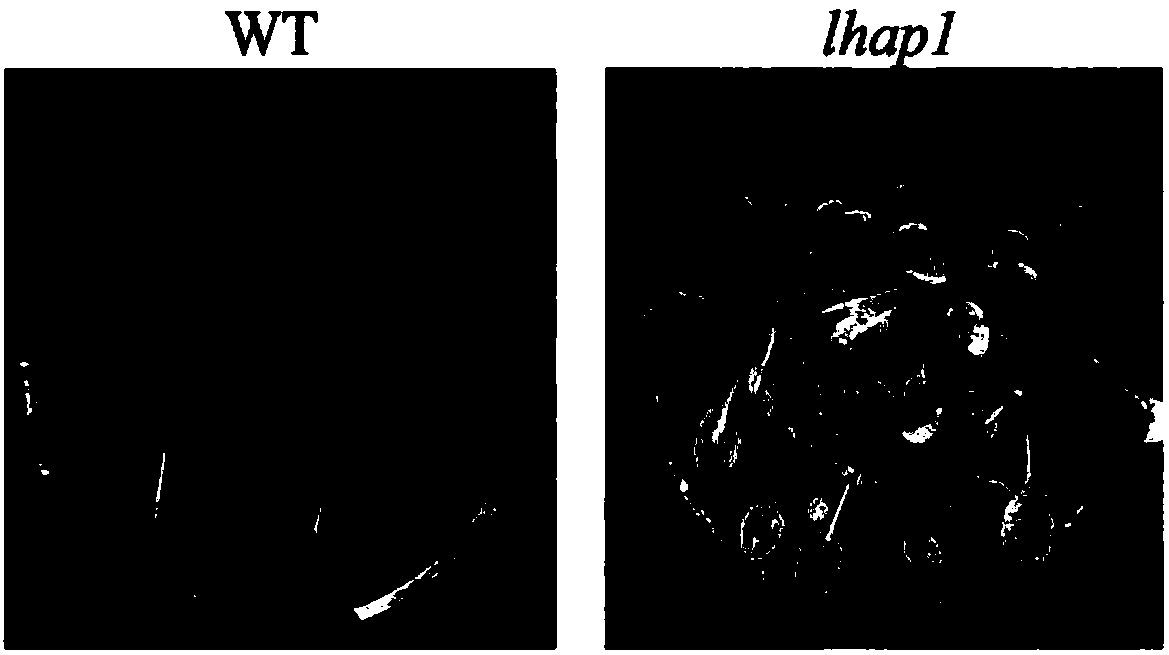
[0076] The seeds of wild-type Arabidopsis and mutants (T-DNA insertion mutants ordered from the NASC germplasm resource bank, the seed number is SALK_031802C) were treated with low-temperature vernalization (4°C light-proof treatment after soaking in water), and after 72 hours of treatment Sterilize with 10% sodium hypochlorite for 10 minutes, wash with sterile water 4 times, sow on MS medium containing 2% sucrose and 0.8% agar, place at 22°C, light intensity 100umolm -2 the s -1 and a photoperiod of 12h / 12h (light / dark) were cultivated to obtain wild-type Arabidopsis seedlings (WT) and mutant plants (lhap1).
[0077] The results showed that: compared with the wild-type plants, the mutant plants (lhap1) germinated later than the wild-type plants, and grew slowly. The leaf yellowing phenotype was aggravated during drought ( figure 1 ).
[0078] ...
Embodiment 2
[0102] Embodiment 2, acquisition of complementary plants and analysis of photosynthetic function
[0103] 1. Acquisition of Complementary Plants
[0104] 1. Construction of complementary vectors
[0105] Insert the DNA molecule shown in Sequence 1 in the sequence listing between the BamHI and KpnI restriction sites of the pSN1301 vector (purchased from BioVector Plasmid Vector Strain Cell Gene Collection Center, the product number is Biovector105802), and keep other sequences of the pSN1301 vector unchanged , to obtain the recombinant vector.
[0106] 2. Acquisition of recombinant bacteria
[0107] The recombinant vector obtained in step 1 was transformed into EHA105 Agrobacterium (purchased from Beijing Zhuangmeng International Biogene Technology Co., Ltd., product catalog number is ZC142) to obtain recombinant bacteria.
[0108] 3. Conversion
[0109] The homozygous mutant plants obtained in Step 1 of Example 1 were infected with the recombinant bacteria in Step 2 by inf...
Embodiment 3
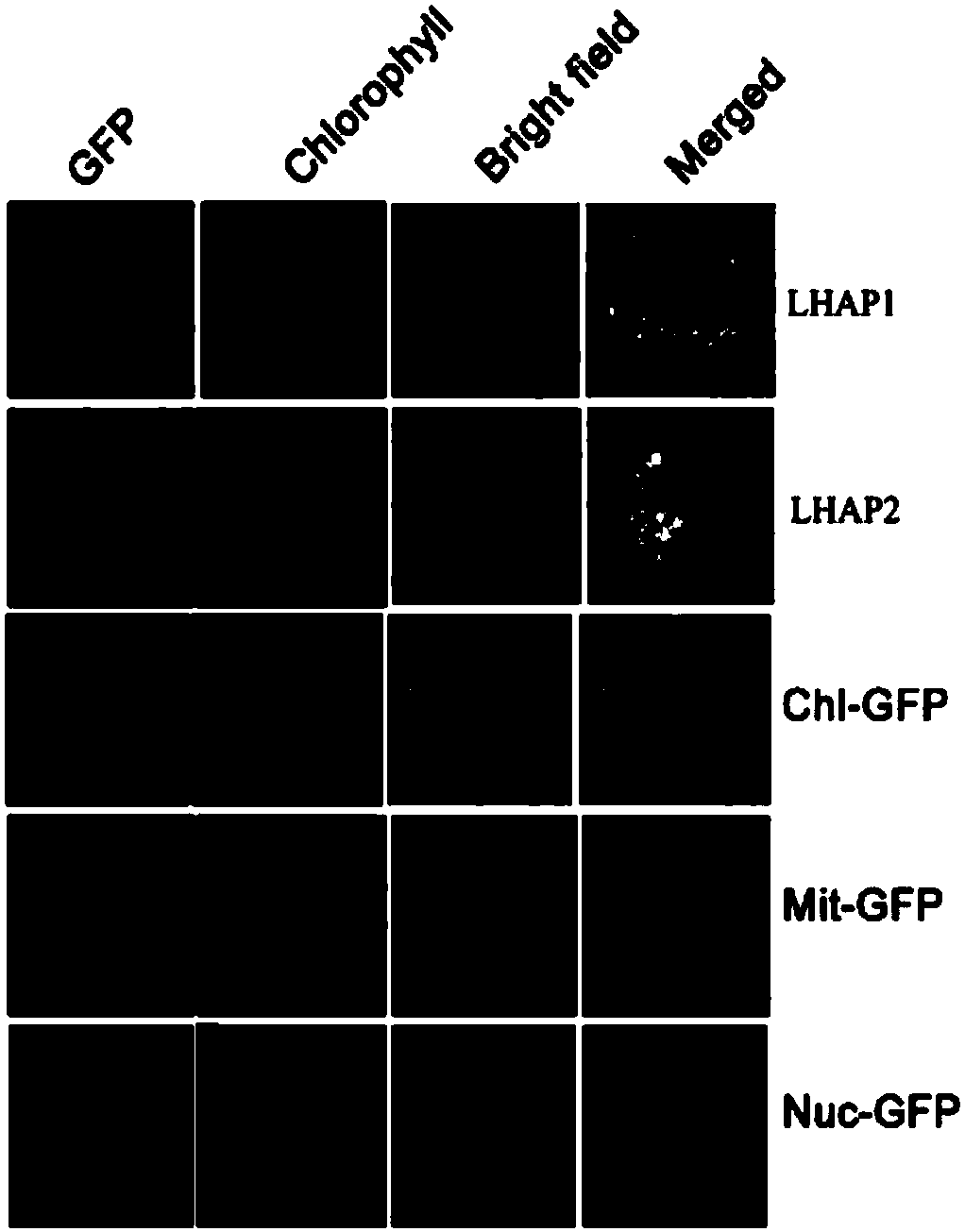
[0135] Example 3. Yeast two-hybrid verification of the interaction between LHAP1 and LHCII transporter LTD
[0136] In this example, through the soluble yeast two-hybrid and other protein interaction experiments, it was found that LHAP1 had a strong interaction with the LHCII transporter LTD.
[0137] 1. Soluble yeast two-hybrid
[0138] Yeast two-hybrid detection system Matchmaker Gold Yeast Two-Hybrid was purchased from Clontech, and the transformation method was completely in accordance with the operation manual. The specific steps are as follows: According to ChlorolP prediction, the first 54 amino acids of the N-terminal of LHAP1 are transit peptides, and the amino acids (Aa55-382) after removing the transit peptide of LHAP1 are divided into the following six segments: LHPA1-1C-terminal (amino acids 55-110), LHPA1-2C-terminal (amino acid 134-147), LHPA1-3C-terminal (amino acid 171-190), LHPA1-4C-terminal (amino acid 214-253), LHPA1-5C-terminal (amino acid 277-313) and LH...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com