Eucaryon alternative splicing analysis method and system based on RNA-seq data
A variable and data technology, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] [Example 1] Basic Analysis
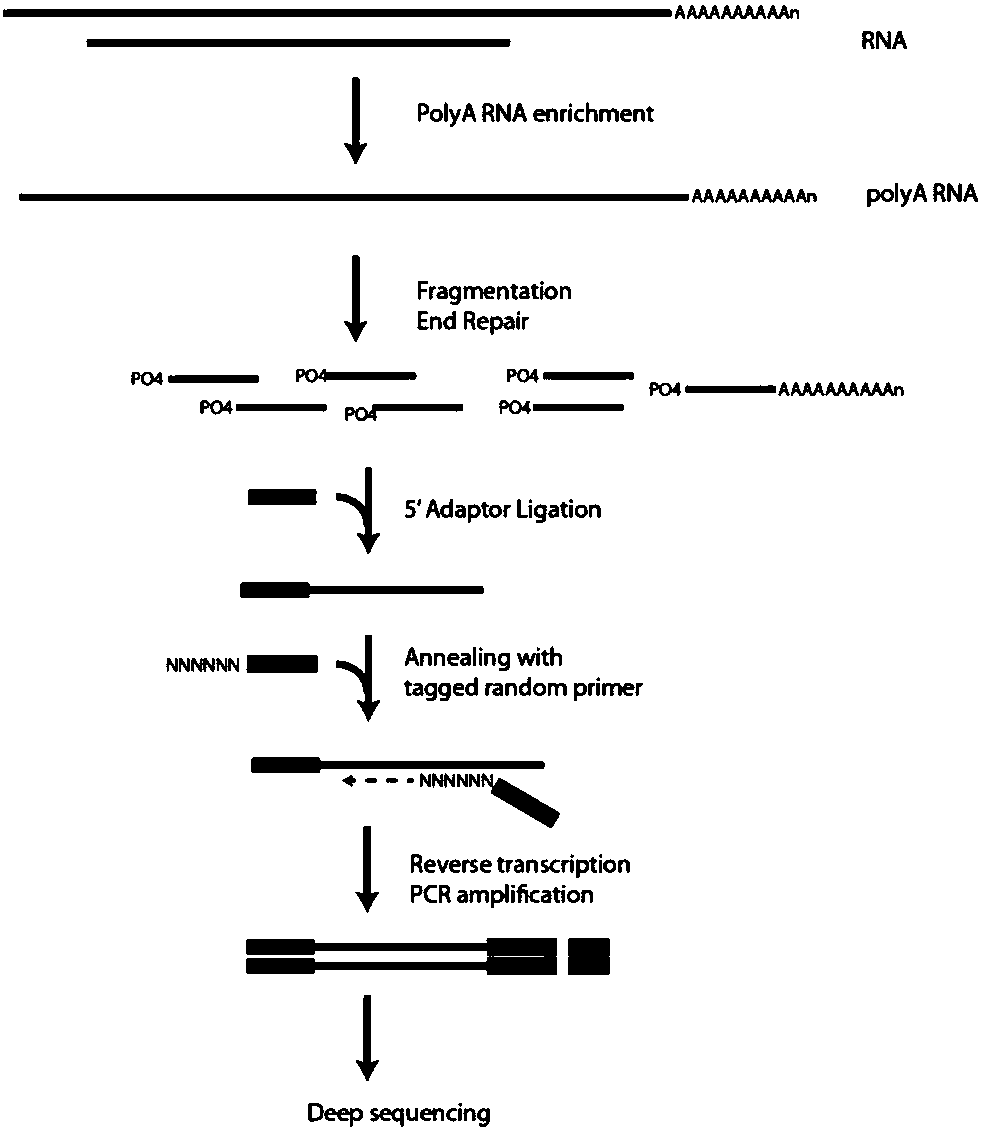
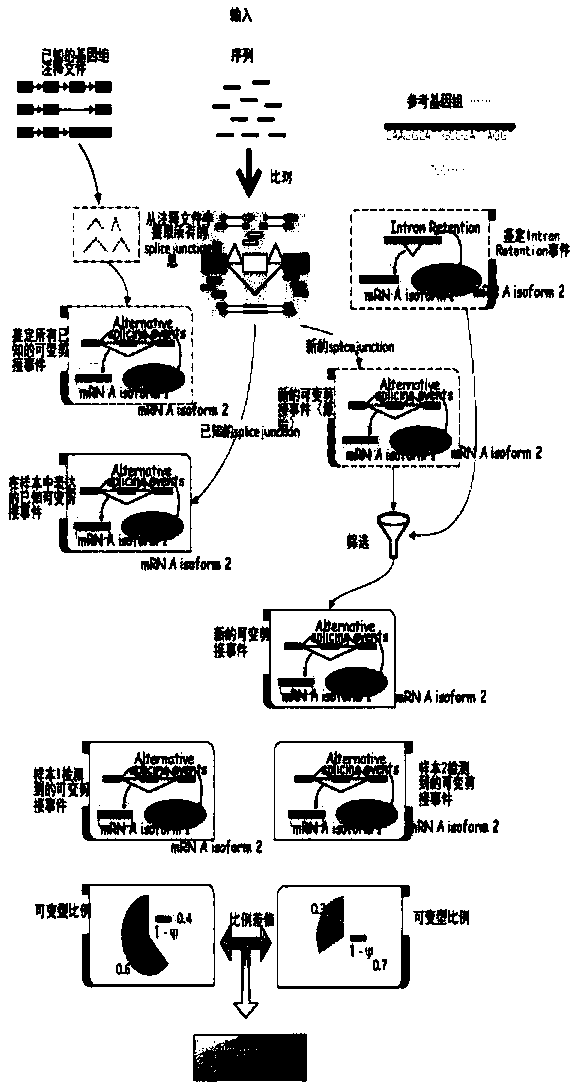
[0051] The present invention uses the illumina Nextseq500 sequencing platform to perform transcriptome sequencing process schematically as follows figure 1 . After obtaining the transcriptome sequencing data of the sample based on the NextSeq500 platform, search for the reference database of the sample and the corresponding annotation files (genes and genomes of the species itself), and then use such as figure 2 The comparative analysis workflow shown provides a detailed analysis of the data. Since all the following processes are based on reference sequences, it is very important to select an appropriate reference database (such as genome sequences and cDNA sequences of public databases such as NCBI and UCSC).
[0052] Data filtering, since some original sequencing sequences have adapter sequences or contain a small amount of low-quality sequences, a series of data filtering is first required to remove impurity data. The data obtained aft...
Embodiment 2
[0068] [Example 2] Alternative splicing analysis
[0069] Algorithms for identification of various alternative splicing events (e.g. image 3 ):
[0070] 1. Gene model:
[0071] Alternative splicing is a relative event. An alternative splicing event contains at least two splicing types, and one splicing type is variable relative to the other. Since a gene has more than one transcript in many annotation files, in order to facilitate the study of the relativity of alternative splicing, we will select a transcript from each gene as a reference model, that is, the gene model for our alternative splicing research. This model is considered to have occurred, and if evidence supporting other splicing patterns, such as new splice sites, is detected, it is considered that an alternative splicing event may have occurred here.
[0072] 2. Exon skipping event (ES):
[0073] There are N exons in the gene model, N>=3, if there is a splice junction between exon [0] and exon [N-1], exon [1...
Embodiment 3
[0106] Figure 8 It is a block diagram of an embodiment of an alternative splicing analysis system based on RNA-seq data of the present invention. Such as Figure 8 As shown, the analysis system includes a data processing module 11, which is used to filter unqualified sequences in the raw sequencing data of each sample, and obtains clean reads of each transcriptome; a comparison module 12, connected to the data processing module 11, used to combine each The transcriptome data is compared to the reference genome, and the unique comparison sequence is extracted; the expression analysis module 13 is connected to the comparison module 12, and is used to calculate the expression RPKM value of the gene; the differential gene analysis module 14, and the expression analysis module Module 13 is connected to screen genes with significant expression differences; functional annotation analysis module 15 is connected to differential gene analysis module 14 and alternative splicing event d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com