Neoantigen Activity Prediction and Ranking Method Based on Tumor Neoantigen Characteristic Value
A eigenvalue, antigen technology, applied in genomics, biological testing, instruments, etc., can solve the problem of low tumor neoantigen screening efficiency, and achieve the effect of efficient and accurate tumor neoantigen screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
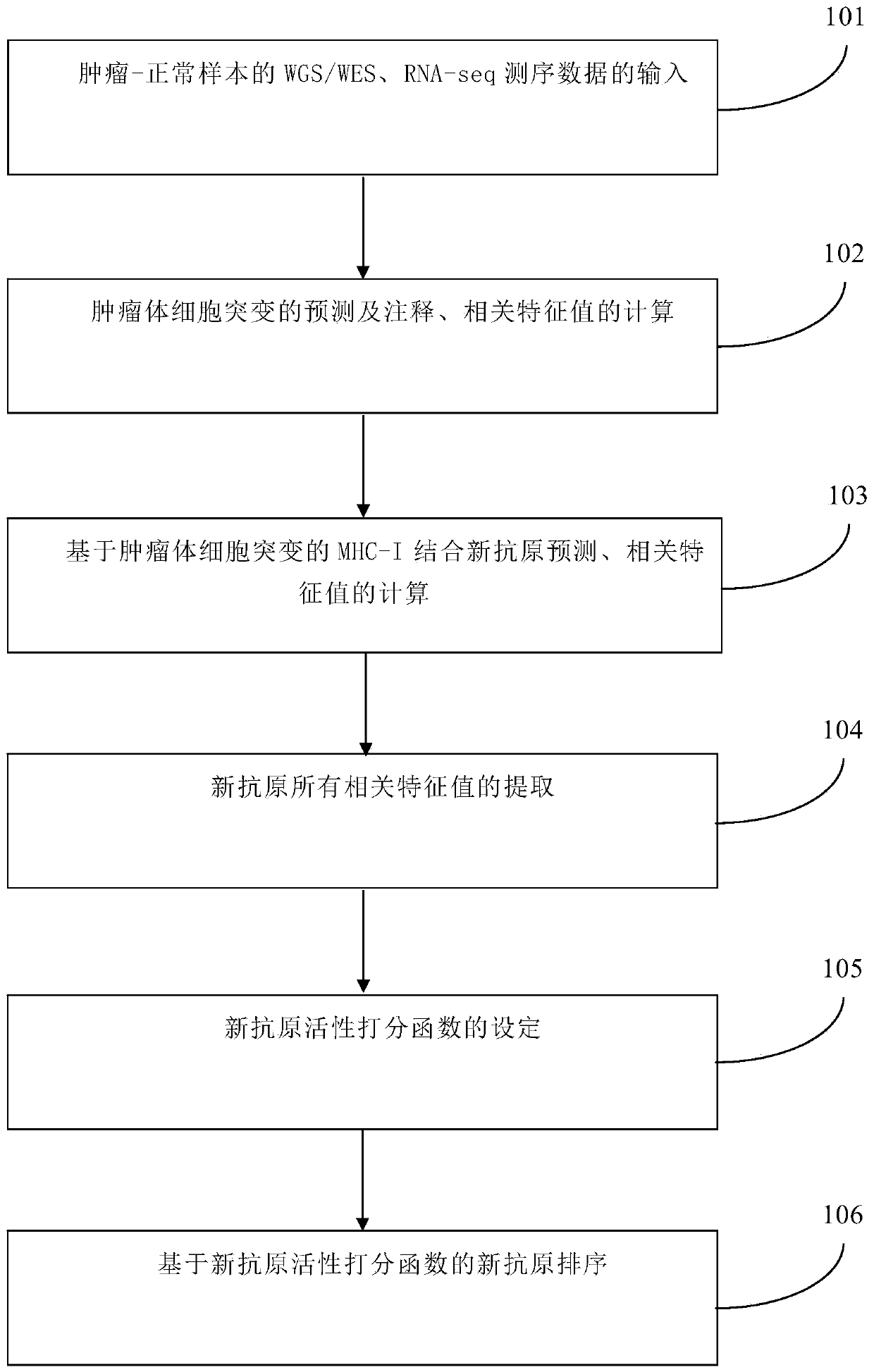
[0039] Such as figure 1 As shown, a neoantigen activity method and sorting method based on tumor neoantigen characteristic value, comprising the following steps:
[0040] Step 101: Input of WGS / WES and RNA-seq sequencing data of tumor-normal samples (using melanoma patient sample-mel_21, Science 2015: Carreno B M, Magrini V, Beckerhapak M, et al.Cancerimmunotherapy.A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells.[J].Science,2015,348(6236):803-8.)
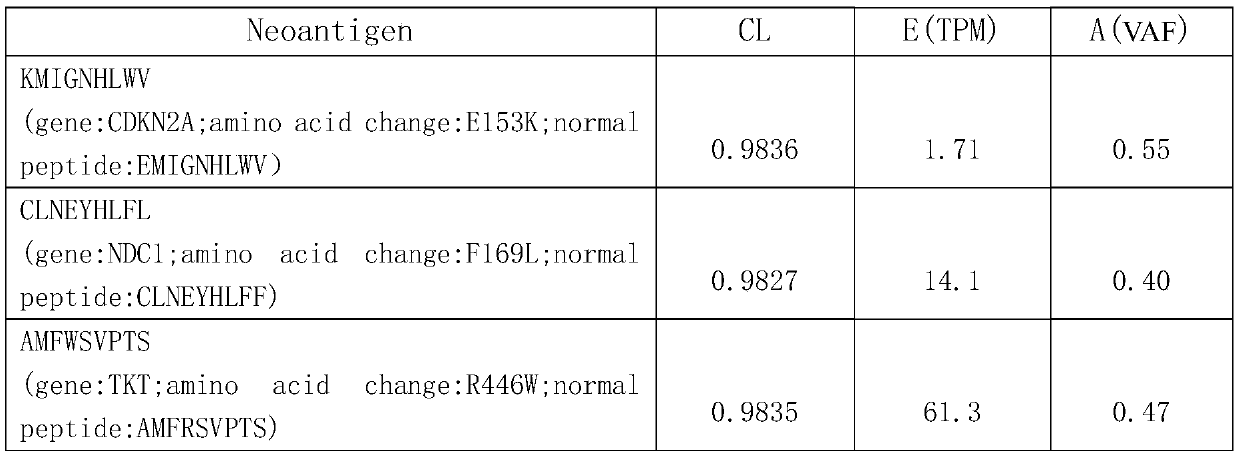
[0041] Step 102: Prediction and annotation of tumor somatic mutations, and calculation of related feature values: based on WGS / WES and RNA-seq sequencing data of tumor-normal samples, use tools such as Varscan / Mutect to analyze and calculate tumor somatic mutations, and call VEP The (Variant Effect Prediction) tool completes mutation annotation, and uses PyClone, Kallisto, and Varscan / Mutect tools to calculate the following feature values: mutant gene clone ratio CL, mutant gene ...
Embodiment 2
[0055] Such as figure 1 As shown, a neoantigen activity method and sorting method based on tumor neoantigen characteristic value, comprising the following steps:
[0056] Step 101: Input of WGS / WES and RNA-seq sequencing data of tumor-normal samples (using melanoma patient sample 2 mel_38, Science 2015: Carreno B M, Magrini V, Beckerhapak M, et al.Cancerimmunotherapy.A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells.[J].Science,2015,348(6236):803-8.)
[0057] Step 102: Prediction and annotation of tumor somatic mutations, and calculation of related feature values: based on WGS / WES and RNA-seq sequencing data of tumor-normal samples, use tools such as Varscan / Mutect to analyze and calculate tumor somatic mutations, and call VEP (Variant Effect Prediction) tool completes the mutation annotation, calls PyClone, Kallisto, Varscan / Mutect tools to calculate the following eigenvalues: mutant gene clone ratio CL, mutant gene expressi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com