Method for quickly detecting bifidobacterium based on high-throughput sequencing and application
A bifidobacteria and gene technology, applied in the field of molecular biology, can solve the problems of low resolution, affecting the industrial application of bifidobacteria, and restricting the research of bifidobacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Follow the steps below:
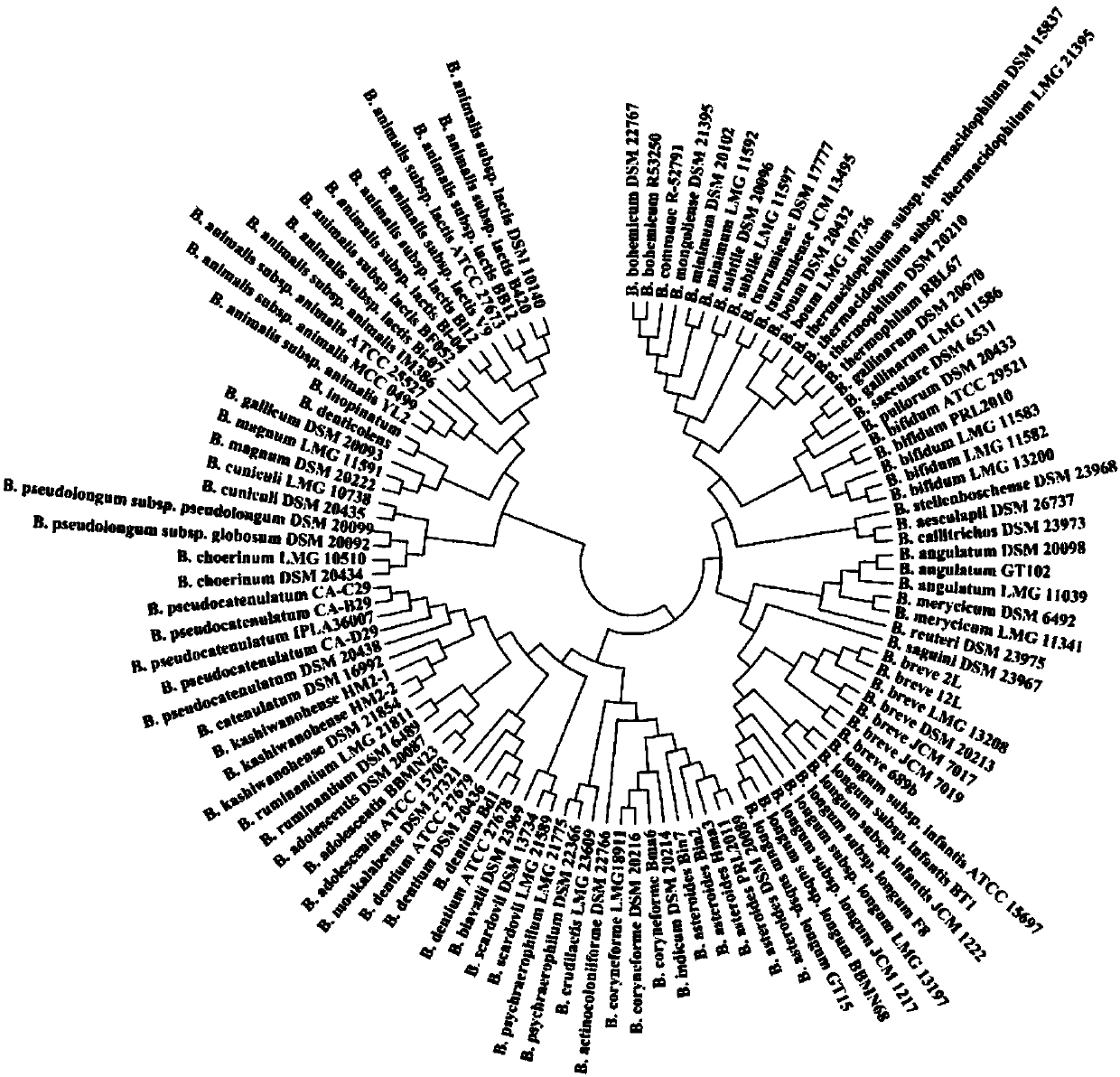
[0038] 1. By downloading 114 groEL gene sequences of different species of Bifidobacterium, construct a phylogenetic tree through MEGA software, such as figure 1 As shown, we can see from the figure that this gene can well distinguish different species of Bifidobacteria, so this gene can be used for identification and detection of different species of Bifidobacteria.
[0039] 2. Construct a groEL gene database, download known groEL gene sequences of bifidobacteria from databases such as NCBI (National Center for Biotechnology Information) and EMBL (European Molecular Biology Laboratory), and use the downloaded sequences to construct a groEL gene comparison database. The constructed groEL gene sequence database is applicable to all known bifidobacteria.
[0040] 3. Adopt the method of high-throughput sequencing to collect the DNA information of the microbial genome of the complex sample to be tested; the extraction of the microbial genome refers...
Embodiment 2
[0045] The detection of different kinds of bifidobacteria in the human feces sample of embodiment 2
[0046] 1. Take a human feces sample, and extract the genomic DNA of the microorganisms in the feces sample, using the DNA extraction kit from MPBiomedicals, USA. The specific operation steps are subject to the operation manual of the kit.
[0047] 2. Design and synthesize Bifidobacterium-specific primer sequences
[0048] Forward primer (SEQ ID NO.1): 5'-TCCGATTACGAYCGYGAGAAGCT-3'
[0049] Reverse primer (SEQ ID NO.2): 5'-CSGCYTCGGTSGTCAGGAACAG-3'
[0050] 3. Use the above-mentioned forward primer and reverse primer, and use the extracted genomic DNA as a template to carry out PCR amplification. The PCR amplification reaction system consists of: genomic DNA template 2ul, Ex taq mix 25ul (TaKaRa), 20uM Each lul of forward primer and reverse primer, add ddH 2 0 to 50ul.
[0051] The reaction conditions of the PCR amplification were: pre-denaturation at 95°C for 5 min, follo...
Embodiment 3
[0055] Determination of content of different species of bifidobacteria in embodiment 3 human feces samples
[0056] 1. According to the method described in steps 1-4 in Example 2, the only difference is that the primer is a general primer for the V3-V4 region of 16S rRNA, which amplifies the V3-V4 region of all microorganisms in human feces, and the database of the V3-V4 region obtained by sequencing The percentage of Bifidobacterium in the total bacterial content was obtained. The specific results are shown in Table 1.
[0057] Table 1 The percentage of Bifidobacteria in the total bacteria in human feces samples
[0058]
[0059] 2. Multiply the percentage of each Bifidobacterium obtained in Step 5 in Example 2 by the content of Bifidobacteria in the stool sample in Step 1 to obtain the percentage of each Bifidobacterium species in human feces . The specific results are shown in Table 2.
[0060] Different species of bifidobacteria account for the percentage of total b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com