Method for determining gastrointestinal tract dysbiosis
A technology for flora imbalance and gastrointestinal tract, which is applied in application, diagnostic recording/measurement, special data processing application, etc., and can solve the problems of relative activity deviation, reduction, and imbalance of microbial groups
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
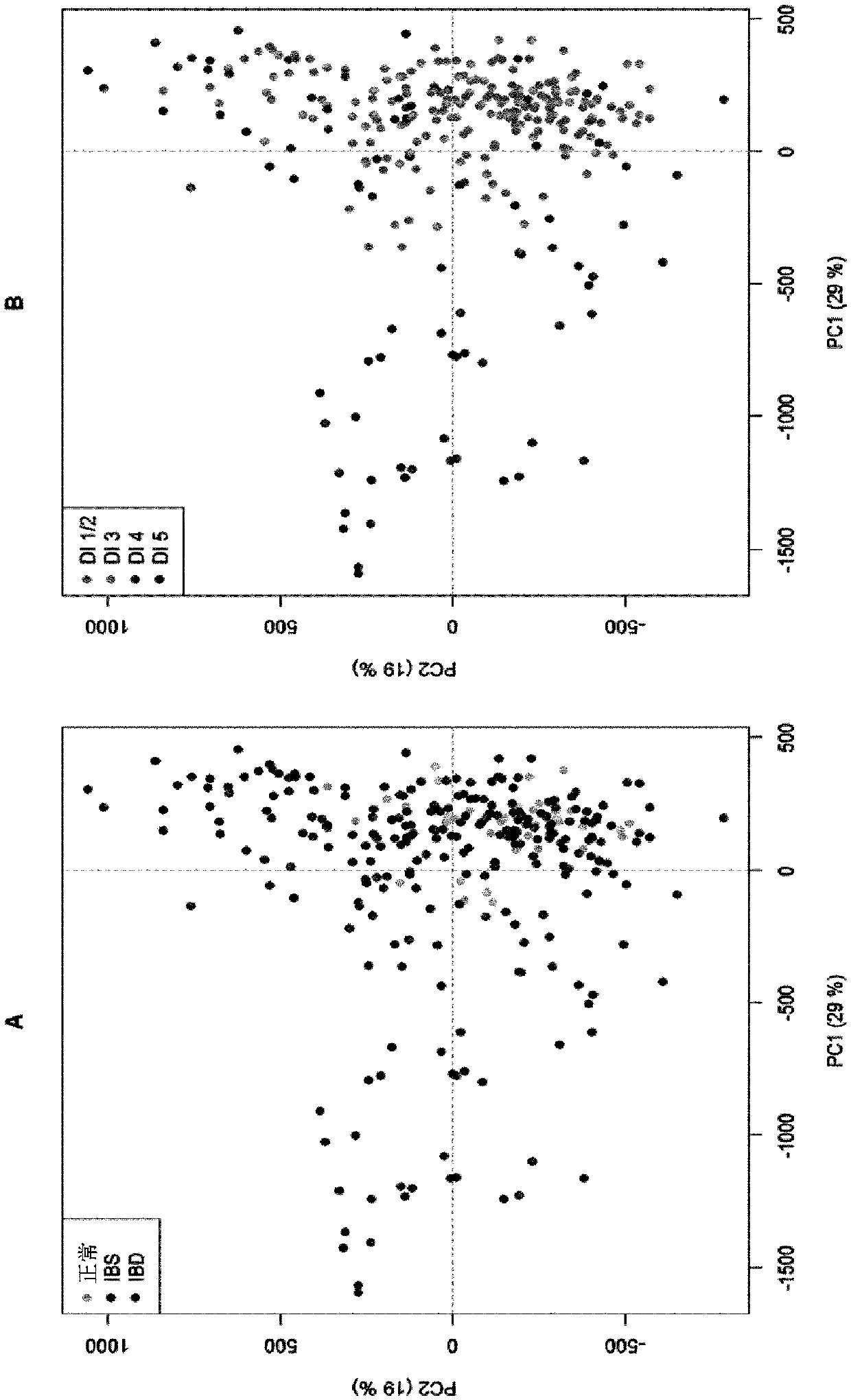
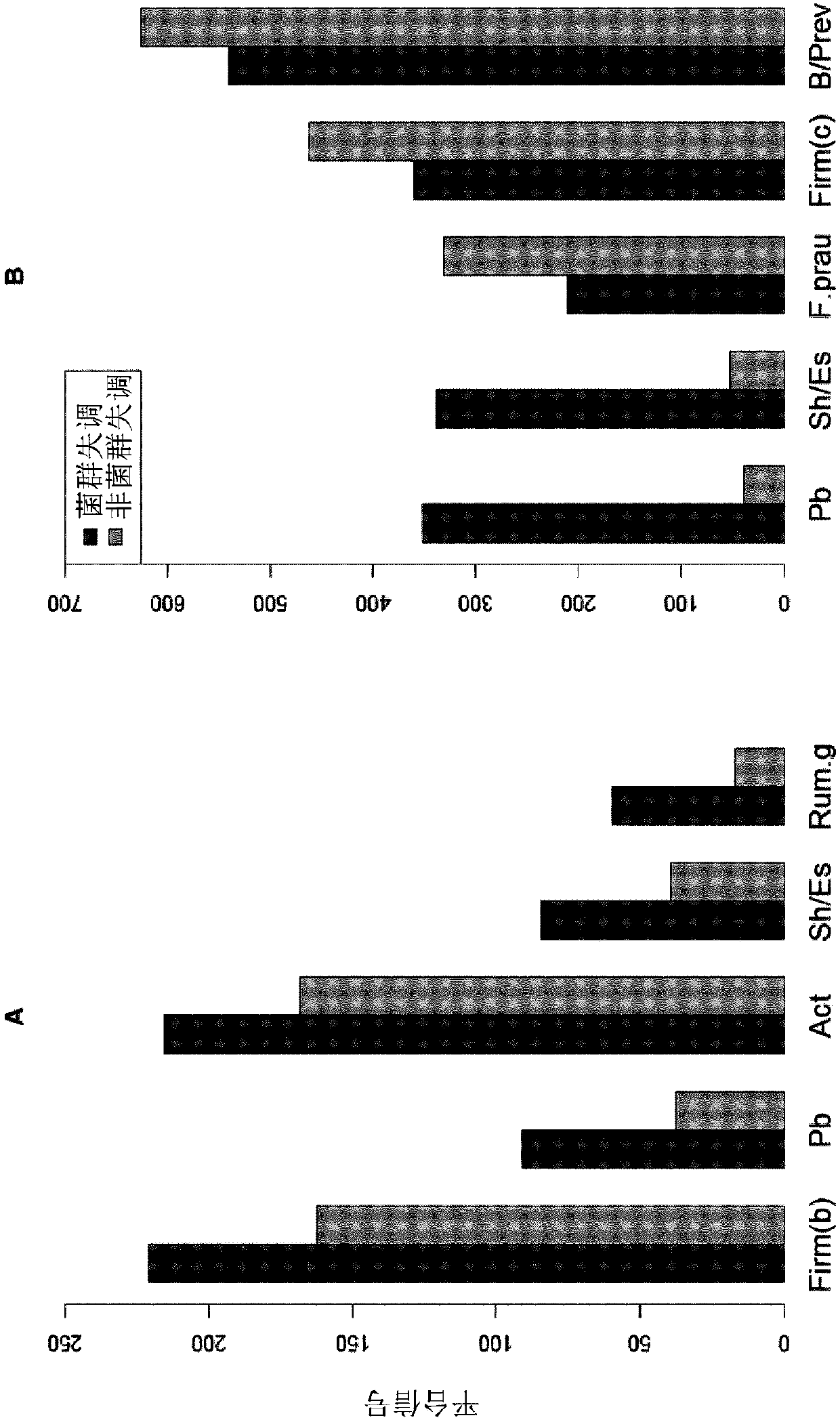
[0145] Example 1 - Targeting and using multiple microbes or groups of microbes to determine dysbiosis in IBS and IBD patients Preparation of 54 probes for gastrointestinal microbiota profile
[0146] Materials and methods
[0147] human sample
[0148] Stool samples were collected from 668 adults (17-76 years; 69% female), including normal individuals (n=297) and patients with IBS (n=236) and IBD (n=135) (Table 1). Fecal samples were collected from hospitals in Norway, Sweden, Denmark and Spain (72%) and workplaces in Oslo, Norway (28%) to address heterogeneity. Normal donors have no clinical signs, symptoms or history of IBD, IBS or other organic gastrointestinal related disorders such as colon cancer. IBS samples were collected as part of a prospective study using Rome III diagnostic criteria to identify IBS. The distribution of IBS subtypes was 44% IBS-diarrhea, 22% IBS-alternating, 17% IBS-constipation, 11% IBS-not subtyped and 4% IBS mixed. The diagnosis of IBD ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com