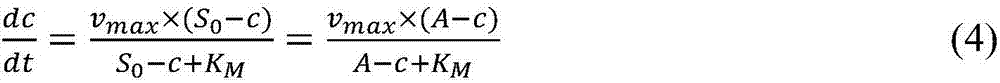Method for screening feature peptide fragment for quantitatively detecting protein in food
A screening method and a technology of characteristic peptides, which are applied in the field of screening of characteristic peptides for quantitative detection of proteins in food
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0100] Example 1: The method for optimizing the quantitative characteristic peptides of bovine serum albumin in raw milk.
[0101] 1. Peptide specificity evaluation: use the BLAST test in proteomics (https: / / blast.ncbi.nlm.nih.gov) to analyze and determine the matching of each theoretical enzymatic peptide in bovine serum albumin Score and number of interferences. Among them, the matching score is determined by the length of the peptide and the matching degree of the database, and the number of interference is the main parameter to determine the specificity of the peptide. Searching in the whole proteomics database, the more the number of interfering peptides that match 100% of the target peptide, the weaker the specificity of the peptide.
[0102]As shown in Table 2, according to the test results of each peptide, 16 candidate target peptides with strong specificity were finally selected, namely LVNELTEFAK, TCVADESHAGCEK, NECFLSHK, LKPDPNTLDEFK, AEFVEVTK, YICDNQDTISSK, SHCIAE...
Embodiment 2
[0131] Example 2: Methodological verification of the quantitative characteristic peptides of bovine serum albumin in raw milk.
[0132] The two peptides LVNELTEFAK and LVVSTQTALA screened in Example 1 were validated in the quantitative experiment of bovine serum albumin in milk.
[0133] Method validation includes four parts: linearity and range, detection limit and quantitation limit, recovery rate and precision.
[0134] According to the method verification results, the linearity of the two quantitative characteristic peptides is good (R 2 >0.999), the range is 1ppm-100ppm; the limit of detection is 0.21-0.22mg / kg, the limit of quantification is 0.70-0.74mg / kg; the recovery rate is 99.0%-111.1%, the precision is 2.20%-6.67%, in line with ISO / IEC 17025:2005 standard.
Embodiment 3
[0135] Example 3: Method for optimizing the quantitative characteristic peptides of α-lactalbumin in infant formula powder.
[0136] Refer to Example 1 for the preferred method of characteristic peptides, and the results are shown in Table 3.
[0137] Table 3 Characteristic parameters of quantitative characteristic peptides of α-lactalbumin
[0138]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com