sgRNA for promoting development of cotton lateral root and application thereof
A cotton and lateral root technology, applied in the field of plant genetic engineering, can solve problems such as inability to effectively verify sgRNA activity, difficulty in cotton protoplasts, and no cotton found, so as to improve stress resistance, increase root surface area, and improve absorption and utilization. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0065] Example 1 Obtaining of Cotton Arginase (ARG) Gene GhARG Target Sequence
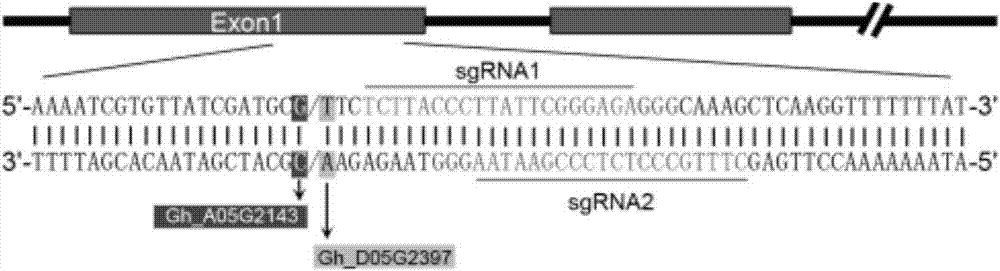
[0066] Use the NCBI online database to search for the cotton arginase gene GhARG and download it. According to the sgRNA design method of Wagner JC et al. The target sequence was designed for each exon sequence, and optimized according to the target site, the number of mismatched bases, and the mismatched position, and a total of two target sequences were obtained, which were named sgRNA1 and sgRNA2 ( figure 1 ).
[0067] sgRNA1: 5'-TCTTACCCTTATTCGGGAGA-3' (SEQ ID No.1);
[0068] sgRNA2: 5'-CTTTGCCCTCTCCCGAATAA-3' (SEQ ID No. 2).
Embodiment 2
[0069] Example 2, CRISPR / Cas9 gene editing system expression vector construction
[0070] Proceed as follows:
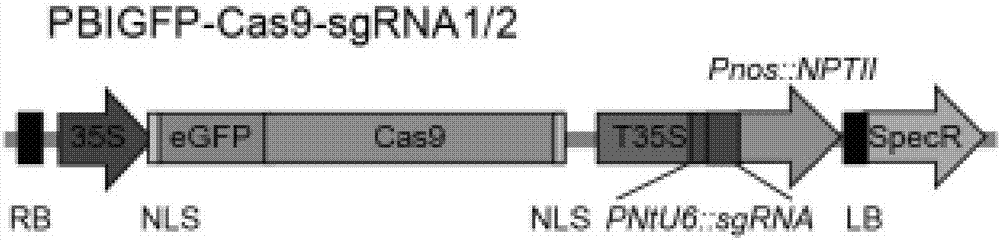
[0071] (1) The tobacco NtU6 promoter was added to the 3' end of the sgRNA1 sequence obtained in Example 1, and the hairpin structure and terminator sequence combined with the Cas9 enzyme were added to the 5' end to form a complete expression cassette. According to the enzyme cutting site of the vector pBI121, an additional 4 bases TCGA was introduced at the 5' end of the expression cassette to obtain sequence 3 (see SEQ ID No.3), and sequence 3 (SEQ ID No.3) was reversely complementary to obtain For the reverse sequence of sgRNA1, 4 bases CTAG were introduced at the 5' end of the reverse sequence to obtain sequence 4 (see SEQID No.4).
[0072] Sequence 3:
[0073] 5'-TCGACATAGCGATTGTCTTACCCTTATTCGGGAGAGTTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT-3' (SEQ ID No. 3)
[0074] Sequence 4:
[0075] 5'-CTAGAAAAAAAGCACCGACTCGGTGCCA...
Embodiment 3
[0092] Embodiment 3, the acquisition of cotton transgenic plant
[0093] Including the following steps:
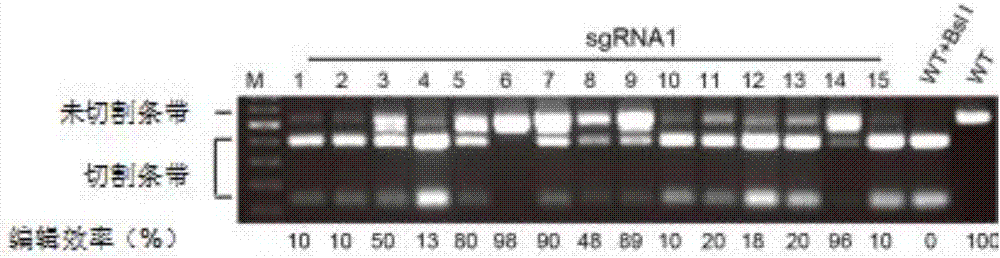
[0094] 1. The expression vector pBIGFP-Cas9-sgRNA1 obtained in Example 2 was introduced into Agrobacterium GV3101 (Agrobacterium GV3101 was provided by the Institute of Biotechnology, Chinese Academy of Agricultural Sciences) to obtain Agrobacterium containing the gene editing expression vector. The specific steps are as follows: Add 10 ng of the expression vector pBIGFP-Cas9-sgRNA1 obtained in Example 1 into 100 μL of Agrobacterium GV3101 competent, place on ice for 30 minutes, freeze in liquid nitrogen for 10 minutes, heat shock at 42°C for 90 seconds, and place on ice for 3 minutes; add 700 μL of liquid YEB medium was cultured on a shaker at 28°C and 200 rpm for 6 hours to recover the bacteria. Use a pipette to insert onto solid YEB medium containing spectinomycin and kanamycin resistance, and culture at 28°C for 2 days. A single colony was picked and cultured in 500u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com