Method for screening interacting protein based on bimolecular fluorescence complementation technique
A technology for green fluorescent protein and protein, applied in the field of screening interacting proteins, can solve the problems of high cost, heavy workload and high fluorescence background value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1, the establishment of the BiFC system based on YGFP
[0065] 1. Transformation of yeast expression vectors pPC86 and pDBLeu
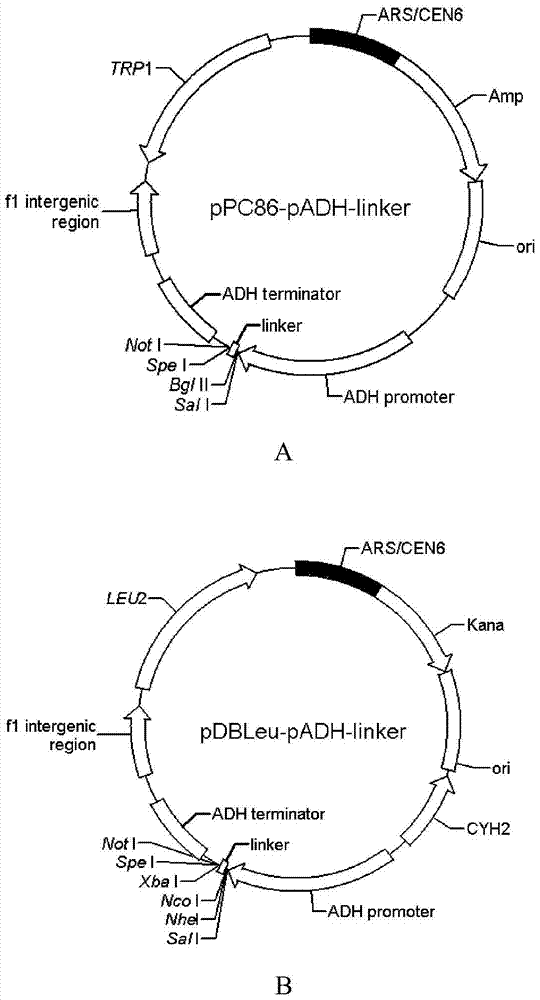
[0066] 1. Transform yeast expression vector pPC86 and construct pPC86-pADH-linker
[0067] (1) Restriction endonucleases KpnI and SpeI were used to double-digest the yeast expression vector pPC86, remove the ADH promoter-NLS-GAL4AD sequence, and retain a vector backbone fragment with a size of about 5209bp.
[0068] (2) Using pPC86 as a template, using primers pPC86-pADH-F and pPC86-linker-R to carry out PCR amplification, the ADH promoter-linker sequence was amplified, and it was treated with KpnI and SpeI double enzyme digestion and then connected into the step ( 1) The backbone fragment of the pPC86 vector that has been digested with KpnI and SpeI to form the pPC86-pADH-linker vector.
[0069] Upstream primer pPC86-pADH-F:
[0070] 5'-AAAGGTACCATCCGGGATCGAAGAAATG-3';
[0071] Downstream primer pPC86-linker-R:
[0072] 5'-CTAG...
Embodiment 2
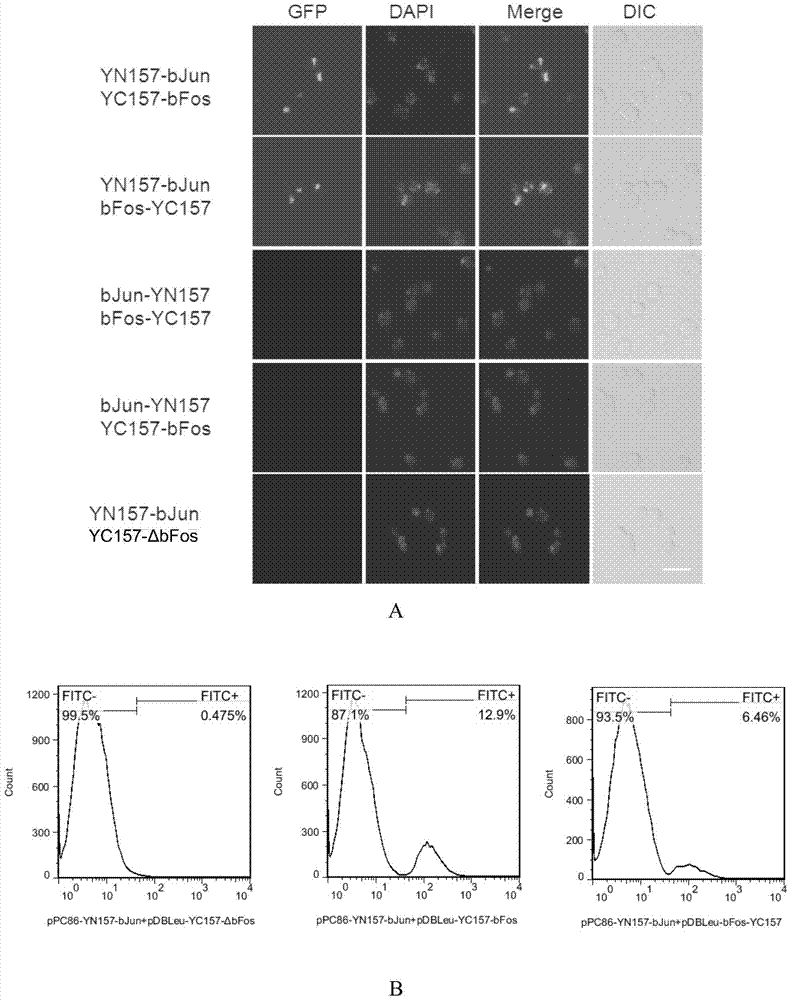
[0089] Example 2, using the BiFC system established in Example 1 to verify the interaction between bJun protein and bFos protein
[0090] It is known that bJun protein and bFos protein are a pair of interacting proteins (see "Hu, C.D., Chinenov, Y. & Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Molecular cell 9,789-798( 2002).”, the present embodiment selects “bJun protein and bFos protein” that interact with each other as the positive test group, “bJun protein and the ΔbFos protein that has lost the interaction domain with bJun protein” (published “bFos protein interacts with The reference of bJun protein interaction domain" is "Hu, C.D., Chinenov, Y.&Kerppola, T.K.Visualization of interactions among bZIP and Refamily proteins in living cells using bimolecular fluorescencecomplementation. Molecular cell 9,789-798(2002).") as In the negative experiment group, the BiFC system esta...
Embodiment 3
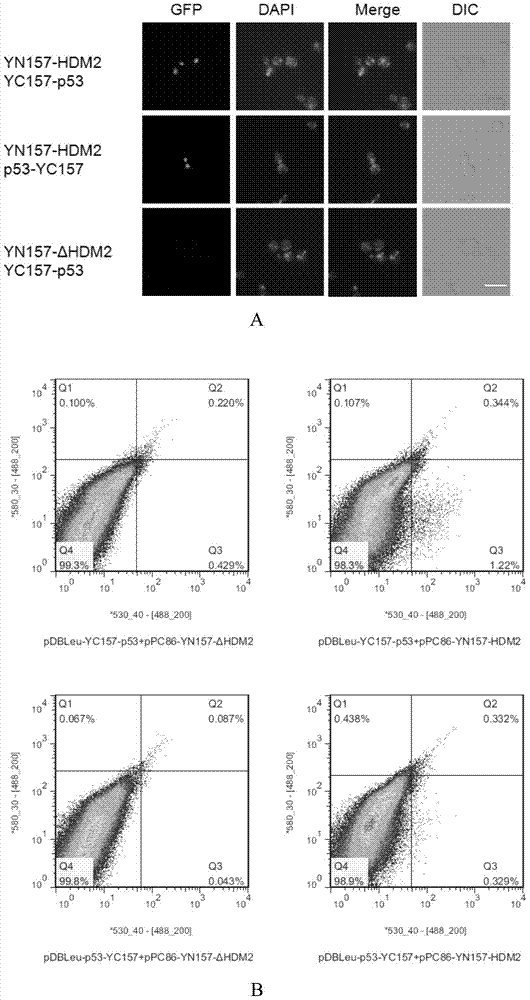
[0145] Example 3, application of the BiFC system established in Example 1 to screen for p53-interacting proteins
[0146] After establishing the YGFP-based BiFC system in yeast, this example uses p53 protein as a bait to screen its interacting proteins from a humanized cDNA library.
[0147] 1. Construction of pDBLEu-YC157-p53 and pDBLEu-p53-YC157 vectors
[0148] 1. Construction of pDBLeu-YC157-p53 vector
[0149] Using the coding gene of p53 protein shown in Sequence 9 in the Sequence Listing as a template, primers were used to perform PCR amplification on p53-F1 and p53-R1 to obtain amplified products with restriction sites XbaI and NotI at both ends.
[0150] p53-F1:5'-ACC TCTAGA ATGGATGATTTGATGCTGTCCC-3';
[0151] p53-R1: 5'-ATTT GCGGCCGC GTCTGAGTCAGGCCCTTCTGT-3'.
[0152] The amplified product was double-digested with restriction endonucleases XbaI and NotI, and after gel recovery, it was ligated with the pDBLeu-YC157 vector (see Example 2 for the specific constru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com