Forest tree genetic evaluation precision improving method using SSR marker information
A technology of marker information and accuracy, applied in the field of forest tree genetics and breeding, can solve the problems of inability to provide pedigree information, pedigree information, inability to accurately estimate tree genetic parameters, and inability to accurately estimate genetic variance, etc., and achieve the effect of improving the effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] The method for accurately evaluating forest tree genetics of the present invention includes the steps of family individual data collection, SSR marker data construction, model establishment and data analysis; details are as follows:
[0049] 1. Clonal individual data collection
[0050] Measure the ground diameter of all potted individuals, and then input the corresponding ground diameter data according to the clone number.
[0051] 2. Construction of SSR labeled data
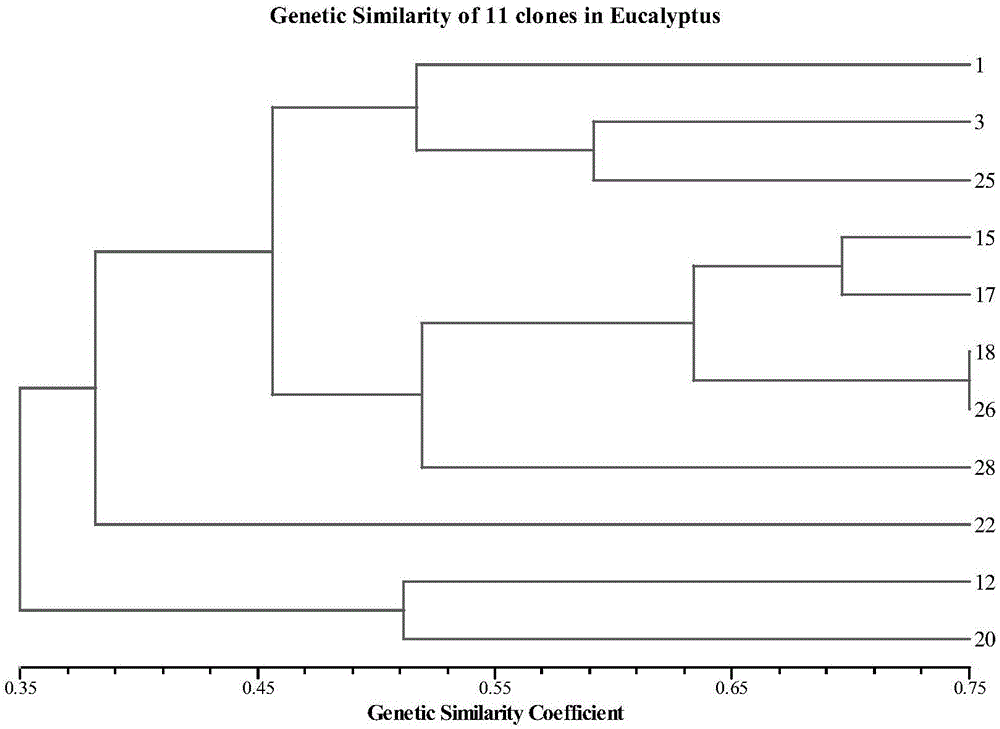
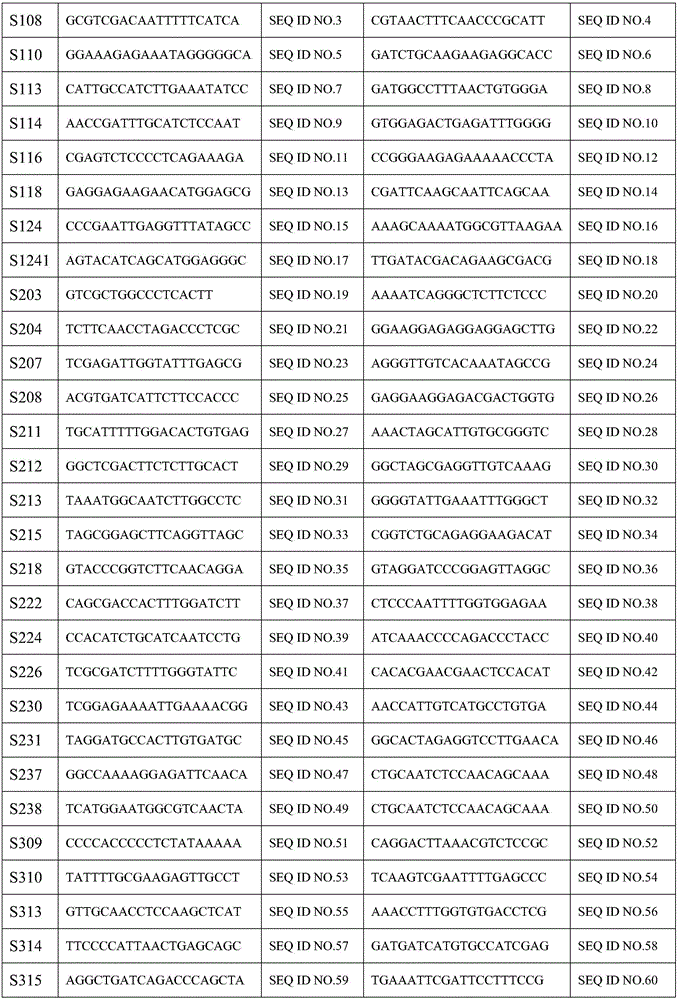
[0052] SSR marker data is a polymorphism data set amplified by clones to different SSR markers and used to calculate the genetic identity between clones. Specific construction method: the DNA of all clones must be extracted, and the amplification of 134 pairs of SSR markers should be completed in sequence. The band was recorded as 1, and the absence of band was recorded as 0. The SSR marker data matrix was established, and the genetic identity coefficient matrix among the clones was calculated by NTsys...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com