Method for detecting MEG3 gene SNP related to cattle growth traits and application thereof
A technology for growth traits and detection methods is applied in the field of detecting MEG3 gene SNPs related to growth and development traits of Chinese cattle breeds, which can solve the problems of growth trait effects and no reports, and achieve the effects of simple operation and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] The present invention will be further described in detail below with reference to the accompanying drawings and embodiments. The embodiments are an explanation of the present invention, not a limitation.
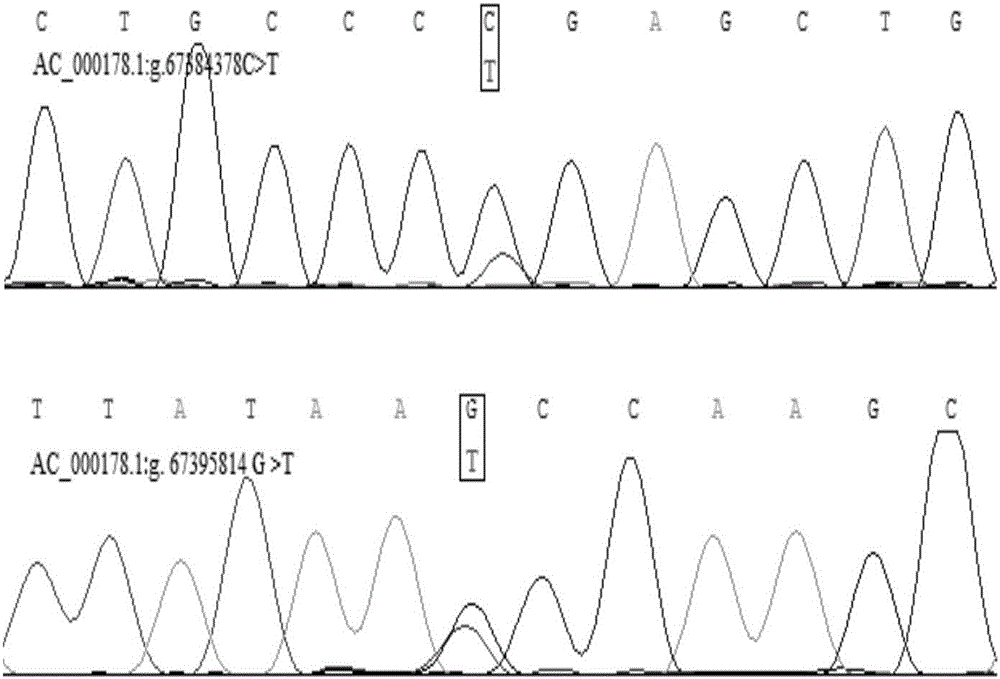
[0027] In the embodiment of the present invention, Chr 21: 67377727-67395997, the candidate region of the bovine MEG3 gene is used as the candidate site:
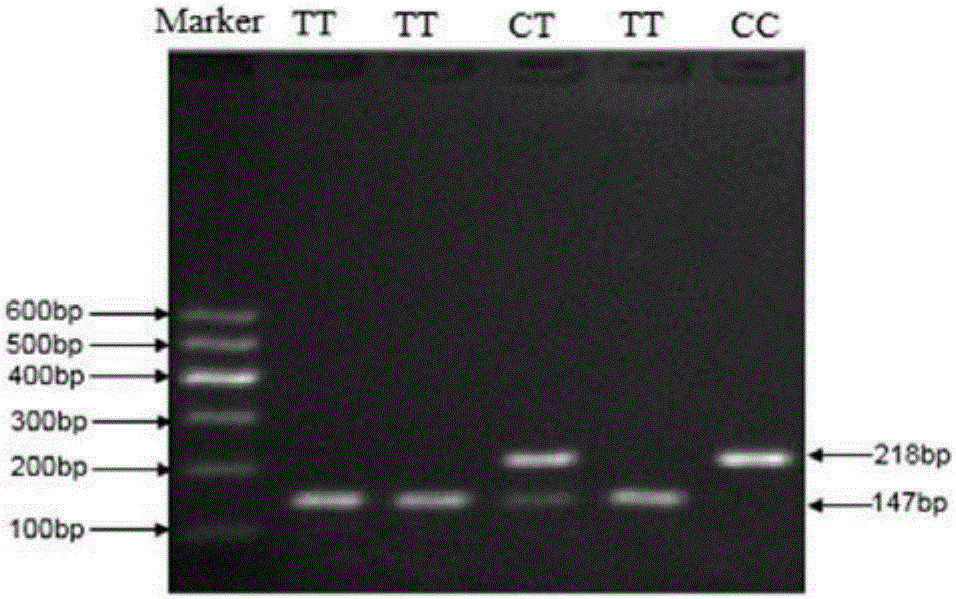
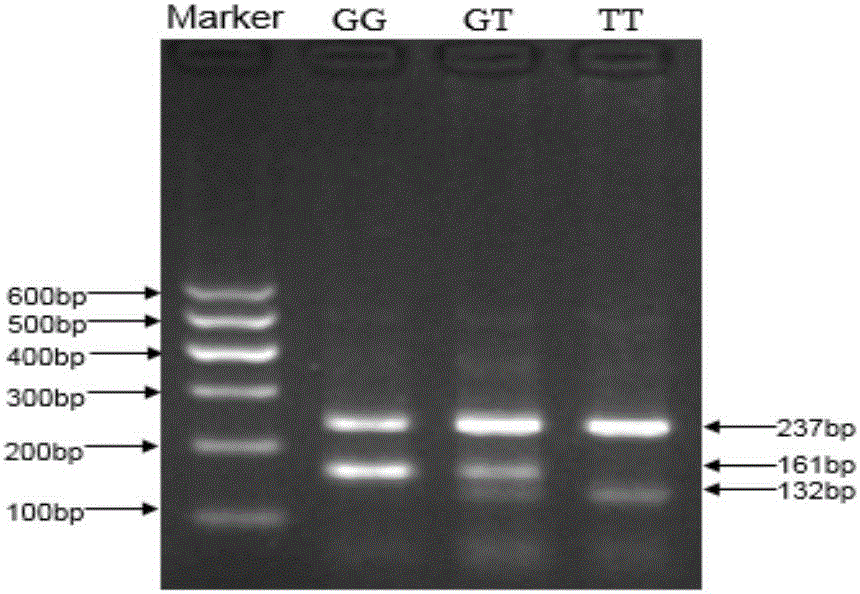
[0028] (1) Using sequencing technology to detect the SNP variation of candidate sites in the cattle population, using PCR-RFLP and Tetra-primer ARMS-PCR technology to classify different SNP sites of different individuals.
[0029] (2) Use SPSS 20.0 software to analyze the association between SNP types and cattle growth traits.
[0030] (3) According to SNP type, select and breed cattle individuals with excellent growth traits.
[0031] 1. Scalper sample collection
[0032] The present invention specifically takes populations of 3 Chinese local cattle breeds, namely Shaanxi Qinchuan cattle (106), Henan Biyang County Xianan cat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com