Specific primers for PCR rapid detection of lactobacillus helveticus and method
A technology based on specific primers and lactobacilli, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection. and other problems, to achieve the effect of fast detection speed, good specificity, and reduced detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
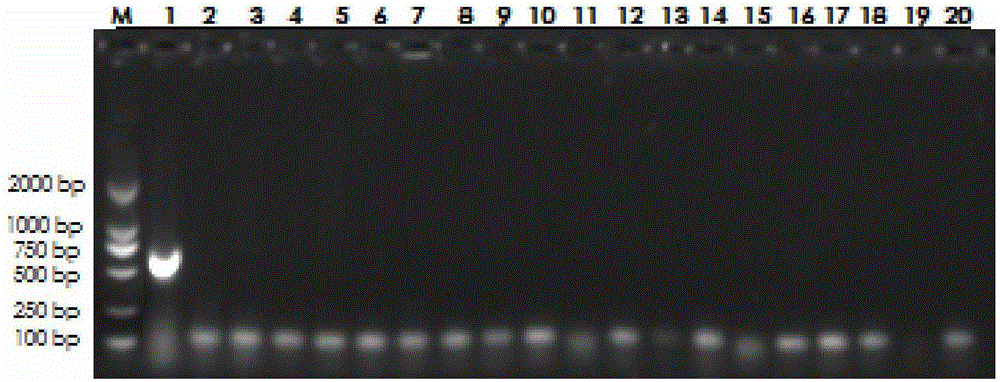
[0027] First, take 1 g of the yogurt sample to be tested by aseptic operation, add it into a sterilized container containing 9 ml of MRS broth, and incubate under anaerobic conditions for 48 hours at 36°C±1°C. Take the cultured bacterial solution and add it to a centrifuge tube, centrifuge at 12000rpm for 1min, discard the supernatant, use bacterial cell precipitation to extract genomic DNA and dilute it for later use.
[0028] Then perform PCR reaction, the total volume of each PCR reaction is 25 μl, including extracted genomic DNA, 2 μl; 10×PCR Buffer, 2.5 μl; Mg 2+ 1.5mmol / L; Taq enzyme 1u; dNTP 0.1mmol / L; Lactobacillus helveticus-specific primers 10pmol each; add ddH at the end 2 0 to 25 μl; set the program parameters of the PCR instrument as follows: denaturation at 95°C for 5min; 30sec at 95°C, 30s at 53°C, and 40sec at 72°C for 35 cycles; extension at 72°C for 5min, and storage at 4°C. Wherein Lactobacillus helveticus-specific primers are:
[0029] L.helv-F: 5'-TTCGAA...
Embodiment 2
[0033] First, take 1 g of the tomato paste sample (containing Lactobacillus helveticus) to be tested aseptically, add it into a sterile container containing 9 ml of MRS broth medium, and incubate under anaerobic conditions at 36°C±1°C for 48h. Take the cultured bacterial solution and add it to a centrifuge tube, centrifuge at 12000rpm for 1min, discard the supernatant, use bacterial cell precipitation to extract genomic DNA and dilute it for later use.
[0034] Then perform PCR reaction, the total volume of each PCR reaction is 25 μl, including extracted genomic DNA, 2 μl; 10×PCR Buffer, 2.5 μl; Mg 2+ Concentration 1.5mmol / L; Taq enzyme 1U; dNTP 0.1mmol / L; Lactobacillus helveticus-specific primers each 10pmol; finally add ddH 2 0 to 25 μl;
[0035] The program parameters of the PCR instrument were set as follows: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 30 sec, annealing at 53°C for 30 s, extension at 72°C for 40 sec, 35 cycles; final extension at 72°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com