Rice early-heading main-effect QTL molecular markers, identifying method thereof, and applications of molecular markers and identifying method
A technology of molecular markers and identification methods, applied in the field of molecular biology, can solve problems such as the inability to apply new varieties of early-maturing rice varieties and differences in regulatory mechanisms, and achieve the effects of facilitating hybridization, speeding up cloning, and enriching knowledge and understanding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1: Screening and positioning of molecular markers
[0050] (1) Construction of early heading chromosome segment substitution line CSSL13 and analysis of substitutional segments
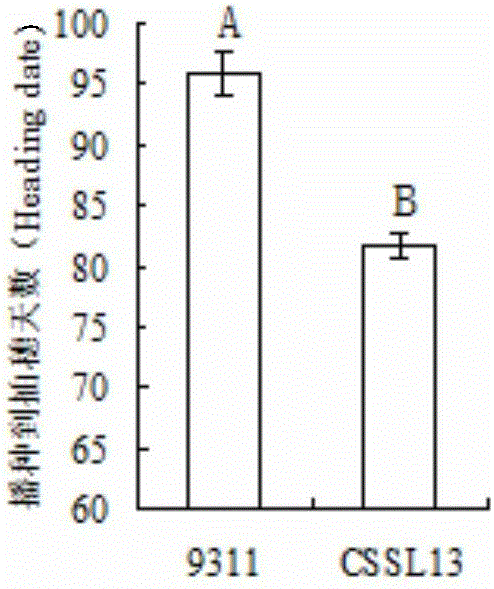
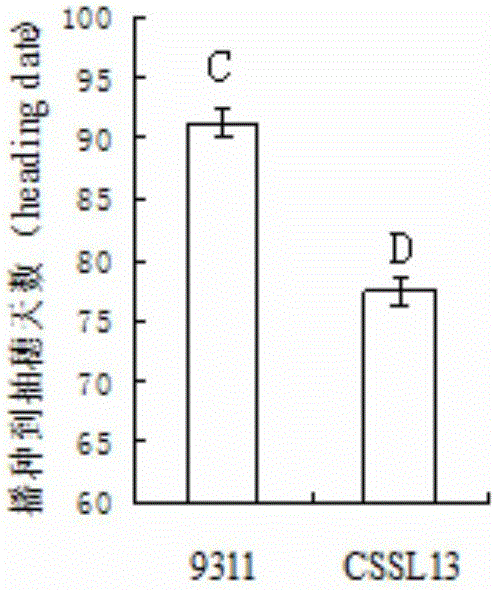
[0051] Using 9311 as the recipient parent and Guangxi common wild rice DP30 as the donor parent, through continuous backcrossing, selfing and directional selection, the BC 4 f 2 The stable genetic early heading chromosome segment substitution line CSSL13 was obtained from this generation.
[0052] According to the SSR marker published on the gramene website (http: / / www.gramene.org / ) and the STS marker designed by the rice indica stalk sequence difference, the polymorphism between the parents DP30 and 9311 was detected, and a total of 408 pairs showing The 408 pairs of polymorphic molecular markers were used to analyze the substitution fragments of CSSL13. The results showed that CSSL13 contained 5 substitution fragments of the donor parent DP30, which were located at the 1st, 2nd, 3rd...
Embodiment 2
[0081] Embodiment 2 Molecular marker identification method
[0082] Using the whole genome DNA of the rice material to be identified as a template, use one or more pairs of primers corresponding to the above-mentioned molecular markers RM6670, M55, M07, M13 and M15 to carry out PCR amplification, and detect the amplified products.
[0083] (1) The donor parent, recipient parent and F were extracted by conventional CTAB method 2 Separate the DNA of each individual plant in the population;
[0084] (2) The reaction system of PCR is as follows:
[0085]
[0086] The PCR reaction conditions were: denaturation at 94°C for 5 min, 30 s at 94°C, 30 s at 56°C, 60 s at 72°C, 35 cycles of amplification, and final extension at 72°C for 10 min.
[0087] The DNA amplification products were detected by polyacrylamide gel electrophoresis, and the amplified DNA bands were recorded by silver staining.
[0088] (3) Result analysis
[0089](1) Use molecular marker RM6670 primers to amplify...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com