Special primers for identification or assisted identification of Propionibacterium acnes, and applications thereof
A P. acnes, auxiliary identification technology, applied in the biological field, can solve the problems of long cycle, cumbersome identification methods, etc., and achieves the effects of high speed, high sensitivity and clear spectrum
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Sample to be tested: Cotton swab after wiping face or post-culture on cotton swab after wiping face
[0033] For extracting the DNA of the sample to be tested, refer to the extraction method of Gram-positive bacteria.
[0034] Specifically, 6 cases of acne patients (ranging from 1 to 4 grades of severity) facial swab samples were taken, and 3 of them were cultured on Columbia blood agar plates, and the cultures were washed and recovered from the plates with PBS buffer, and extracted for culture. total DNA. For the remaining 3 cases, the total DNA of the samples was directly extracted with the QIAamp DNA mini kit (QIAGEN); set aside.
[0035] PCR detection:
[0036] Using the DNA obtained above as templates, primer pair 12 was used for PCR amplification detection. The total volume of the PCR reaction system was 20 μL, including: 0.5 μM upstream primers, 0.5 μM downstream primers, 1 μL template, 10 μL 2×PCR mix (2×PCR Premixed reaction solution), deionized water;
[0...
Embodiment 2
[0042] Sample to be tested: sample culture detection of Propionibacterium acnes and identification of monoclonal bacteria
[0043] Take a cotton swab sample with open comedones (blackheads) on the face and spread it on Columbia blood agar plate for culture. Pick 3 single colonies (2 of which are dry and raised small colonies suspected of Propionibacterium acnes, and the other is a flat translucent colony) for expansion and culture, and set aside.
[0044] PCR detection:
[0045] Using the DAN obtained above as templates, primer pair 34 (see Table 1) was used for PCR amplification detection. The total volume of the PCR reaction system was 20 μL, including: 0.5 μM upstream primer, 0.5 μM downstream primer, 1 μL template, 10 μL 2× PCRmix (2×PCR premixed reaction solution), deionized water;
[0046] The reaction conditions are: pre-denaturation at 94°C for 3 minutes; denaturation at 94°C for 30s, annealing at 60°C for 30s, extension at 72°C for 45s, 35 cycles;
[0047] Electrop...
Embodiment 3
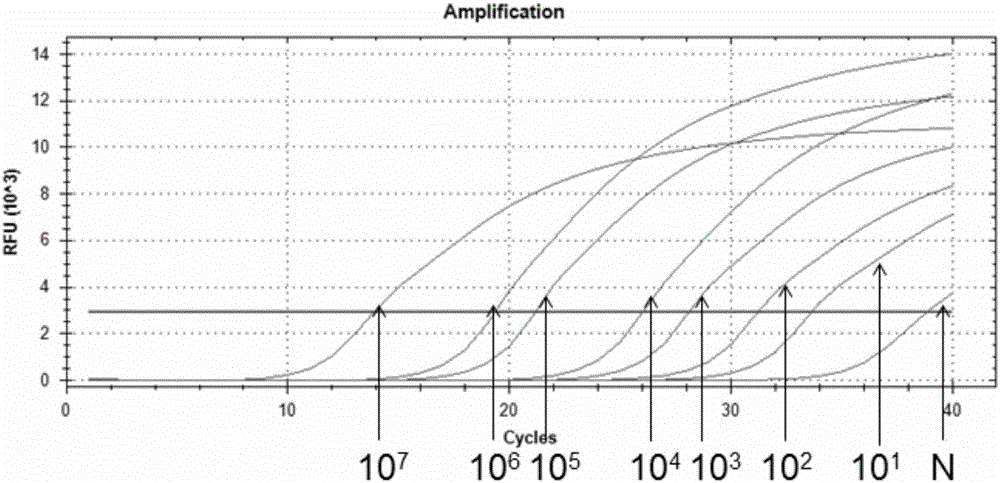
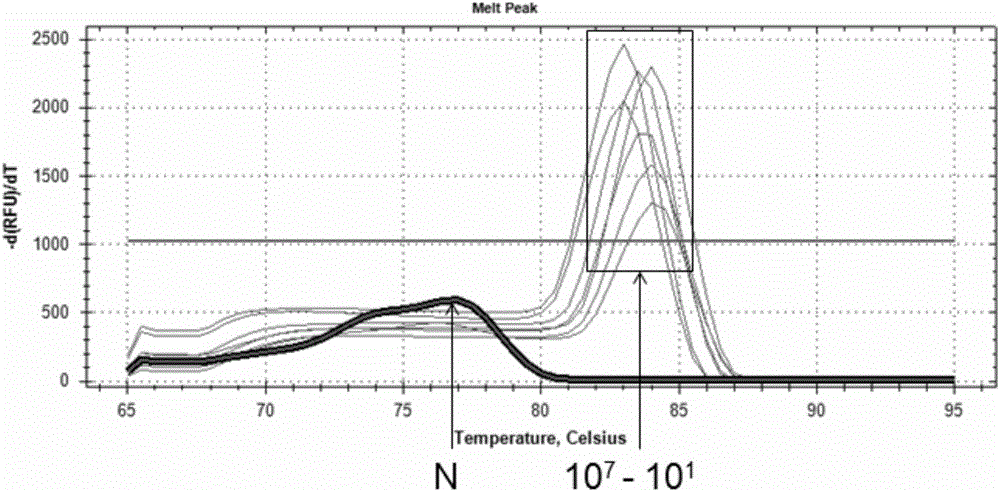
[0051] Sample to be tested: Genomic DNA of a monoclonal culture of Propionibacterium acnes confirmed by sequencing was used as a standard, and its concentration was diluted to 10 7 a / μL; 10 6 a / μL; 10 5 a / μL; 10 4 a / μL; 10 3 a / μL; 10 2 a / μL; 10 1 cells / μL; for use.
[0052] qPCR detection:
[0053] Using the DNA of different concentrations obtained above as a template, PCR amplification detection was performed using primer pair 56. The total volume of the PCR reaction system was 20 μL, including: 0.5 μM upstream primer, 0.5 μM downstream primer, 1 μL template, 10 μL 2×qPCR mix ( 2×qPCR premix reaction solution), deionized water;
[0054] The reaction conditions are: pre-denaturation at 94°C for 3 min; 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s. Fluorescence was detected after each cycle (see Figure 4-7 );
[0055] Depend on Figure 4 Visible, 10 7 for template concentration 10 7 qPCR amplification curve o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com