Isoprene synthase gene and its application
A technology for isoprene and isoprene production, which is applied to isoprene synthase gene and its application field, and can solve the problem of low isoprene efficiency and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: Obtaining of gene fragments
[0029] 1. Extraction of total RNA from leaves of Amorpha fruticosa
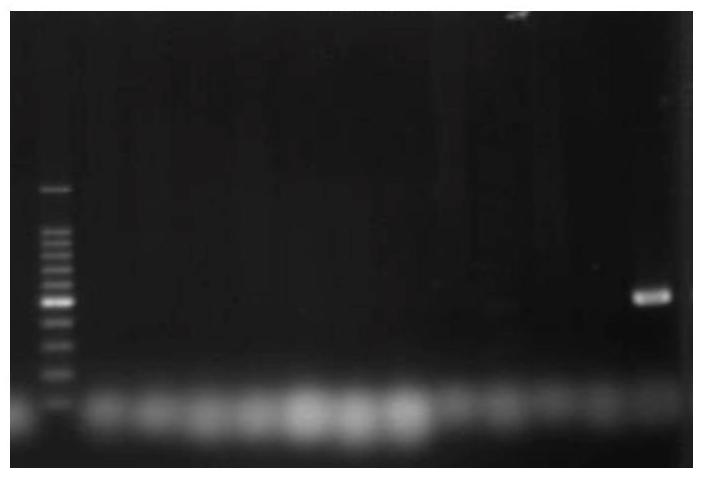
[0030] Collect Amorpha fruticosa leaves, use RNeasy Plant Mini Kit (Qiagen Company) to extract total RNA from Amorpha fruticosa leaves, perform electrophoresis according to the kit instructions ( figure 1 ) to verify the quality of RNA extraction, it can be seen that the integrity of the RNA is good, and subsequent experiments can be performed.
[0031] 2. RT-PCR
[0032] Take Oligo(dT) 20 As a reverse transcription primer, the nucleic acid was reverse transcribed into cDNA according to the reverse transcription kit SuperScript.III First-Strand Synthesis System for RT-PCR (Invitrogen Company) instructions;
[0033] The reaction system is as follows:
[0034] RNA 1 μg
[0035] 10mM dNTP 1μl
[0036] Oligo(dT)20 (0.5μg / μl) 1μl
[0037] 65°C for 5min, place on ice for 1min, add the following 10μl mix
[0038]
[0039] 50°C for 50min, 85°C for 15min, a...
Embodiment 2
[0057] Example 2: Obtaining the full length of the coding region of the AfIspS gene
[0058] The method for obtaining full-length cDNA is SMARTer-RACE, using PCR cDNA Synthesis Kit (Clontech Company) was carried out, and the primers and reagents used below were all except GSP Provided in the PCR cDNA SynthesisKit, follow the kit instructions.
[0059] 1. Preparation of RACE-Ready cDNA
[0060] The reverse transcription system for the first strand of RACE-Ready cDNA is as follows:
[0061]
[0062]
[0063] 2. Design of gene-specific primers:
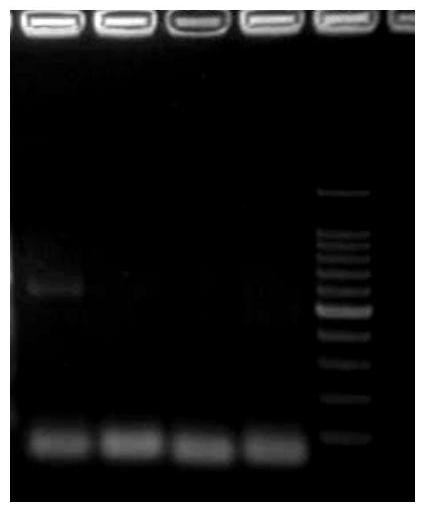
[0064] Design gene-specific primers (GSP) according to the sequence of the obtained AfIspS fragment, use RACE-Ready cDNA as a template, and use GSP and Universal Primer (Universal Primer Mix, UPM) as primers for amplification to obtain 3'-RACEcDNA fragments and 5 '-RACE cDNA fragment. Primer position as Figure 6 As shown, the black part in the middle is the sequence obtained by degenerate PCR, the black part on both sides ...
Embodiment 3
[0087] Embodiment 3: Construction of Escherichia coli isoprene production strain
[0088] The sequences of the full-length primers ZSHFa and ZSHRa are as follows:
[0089] ZSHFa: 5'GTCATGCCATGGGGTGTGCATTGAGCACACAGGATACTC 3'
[0090] ZSHRa: 5' TATCGAGCTCTTCTGCAATTAATTGGAATAGGGTCAAG 3'
[0091] 1. Construction of Escherichia coli expression vector pBAD-AfIspS
[0092] The AfIspS gene fragment obtained using primers ZSHFa and ZSHRa was subjected to NcoI and KpnI double digestion with NcoI and KpnI (TAKARA Company), and the pBAD-HisB expression vector (purchased from Invitrogen Company) was subjected to NcoI and KpnI double digestion, and the AfIspS gene was connected to pBAD- The HisB vector was transformed into trans5α competent cells, and positive clones were selected for sequencing. The nucleotide sequence of pBAD-AfIspS is SEQ ID No.5.
[0093] 2. Construction of isoprene-producing strain MV / pAfispS
[0094] The constructed pBAD-AfIspS and plasmids p1 and p2 were co-trans...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com