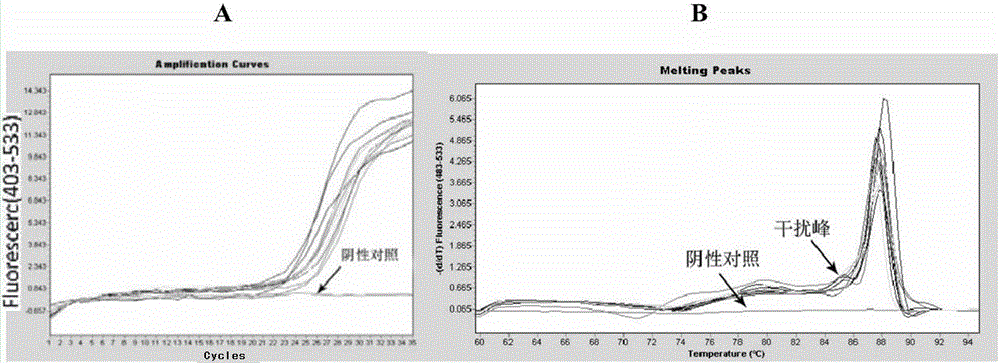
Rapid nucleic acid sequencing method based on fluorescence PCR
A nucleic acid sequencing and sequencing technology, applied in the field of pathogen identification, can solve problems such as routine testing by grassroots testing institutions, lack of systematic genetic markers, and high cost, avoiding biosafety risks, shortening experimental time, and simplifying effect of operation
Inactive Publication Date: 2016-06-22
汕头国际旅行卫生保健中心
View PDF3 Cites 1 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Although the second-generation and third-generation sequencing technologies have been commercialized, the cost is still high, and they are not suitable for daily testing by grass-roots testing institutions
In addition, for the bacterial sequencing identification method, the lack of systematic basic research on related genetic markers is a major reason hindering the large-scale application of this method
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment Construction
[0029] 1 strain of pathogenic bacteria to be identified by sequencing: The strains were isolated from clinical specimens and grew lawns on slant surfaces.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention relates to a rapid nucleic acid sequencing method based on fluorescence PCR, is successfully applied in sequencing of bacterial genes, and can be used for identification of pathogenic bacteria. An FTA card nucleic acid is used for saving pathogenic bacteria nucleic acid, a real-time fluorescence PCR technology is used for amplifying a target fragment, and machine loading is performed for double deoxidization sequencing. A developed fluorescence PCR rapid sequencing reagent kit includes the following reagents: 1) one piece of FTA card; 2) one tube of PCR reaction liquid internally filled with an SYBR GREEN fluorescent PCR reaction solution; 3) one tube of an upstream primer and one tube of a downstream primer which are internally filled with target sequence amplification and sequencing primers; 4) a set of double deoxidization sequencing reagents; 5) a set of sequencing product purification reagents including one tube of a XTerminator solution and one tube of an SAM solution; and 6) one tube of deionized water internally filled with DNA enzyme-free sterile deionized water. Because the FTA card is used for saving the nucleic acid, after the card is dried naturally, the card can be stored for a long time at room temperature, is easy to save, has no need for nucleic acid extraction during PCR amplification, and avoids biological security risks of a bacterial suspension treating operation process during common bacteria gene sequencing.
Description
technical field [0001] The invention relates to a rapid nucleic acid sequencing method based on fluorescent PCR, which is successfully applied in bacterial gene sequencing and can be used for identification of pathogenic bacteria. Background technique [0002] In 1977, Sanger invented the dideoxy DNA sequencing method. So far, the two classic traditional sequencing methods, the dideoxy chain termination method sequencing and the chemical degradation method sequencing, are the most widely used. Completed core technology. The most widely used in this method is the BigDye terminator method of Applied Biosystems. The main steps include DNA extraction, PCR amplification, qualitative detection of PCR products, purification of PCR products, sequencing reaction, purification of sequencing products, capillary electrophoresis, data analysis, sequence splicing (for long fragments), etc. In recent years, due to the improvement and popularization of sequencing technology, as well as th...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12Q1/68
CPCC12Q1/6869C12Q2531/113C12Q2561/113C12Q2563/107
Inventor 蔡颖周霓周广彪陈霖祥魏锐朱俊贤吴烽林昆黄琦容
Owner 汕头国际旅行卫生保健中心
Who we serve
- R&D Engineer
- R&D Manager
- IP Professional
Why Patsnap Eureka
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com