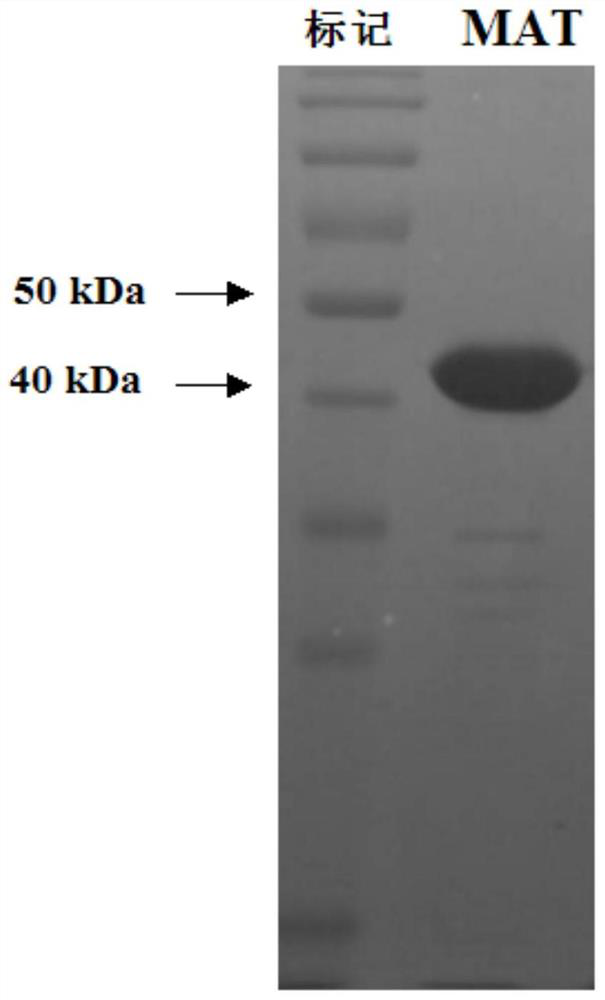
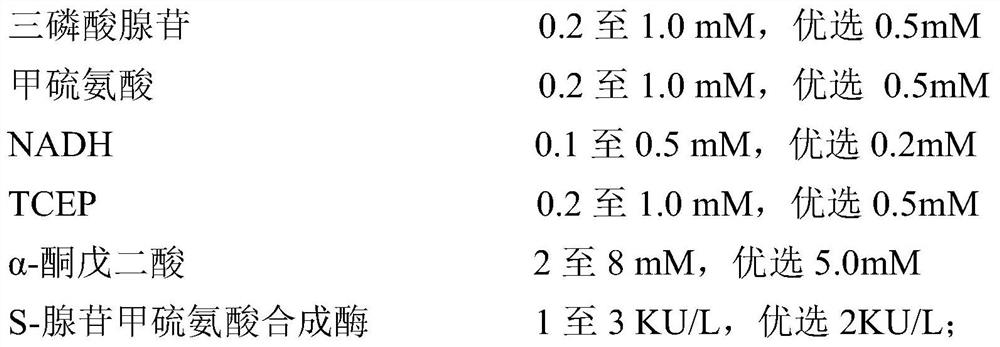
S-adenosylmethionine synthetase preparation, its preparation method and use
A technology of glutamate dehydrogenase and adenosine isotype, applied in the field of biochemistry, can solve problems such as poor stability and interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1: Provide target sequence
[0068] According to the nucleotide sequence of S-adenosylmethionine synthetase published on the NCBI website (Genbank accession number is CP007391.1) is shown in SEQ ID NO:2.
[0069] Design primers, use the Escherichia coli genome as a template, and amplify the gene fragment of S-adenosylmethionine synthetase by PCR;
[0070] The PCR reaction system is as follows:
[0071] wxya 2 O 35μl; 10×Buffer 5μl; dNTP 2μl; template DNA 2μl; forward primer 2.5μl; reverse primer 2.5μl; Taq enzyme 1μl;
[0072] The PCR reaction conditions are as follows:
[0073] 94°C for 3.5min; 94°C for 40s, 55°C for 40s, 72°C for 70s, a total of 35 cycles; 72°C for 10min; 4°C for maintenance;
[0074] Primer sequence:
[0075] Forward: GGGAATTCCATATGATGGCAAAACACCTTTTTAC (SEQ ID No. 3);
[0076] Reverse: ATCCGCTCGAGCTTCAGACCGGCAGCATCGC (SEQ ID No. 4).
[0077] Take the PCR product, and after 1% agarose gel electrophoresis, a product band can be seen at 1...
Embodiment 2
[0079] Embodiment 2: Construction of expression vector
[0080] The PCR product recovered in Example 1 was subjected to double enzyme digestion with NdeI and XhoI enzymes (purchased from New England Biolabs);
[0081] The vector plasmid pET41a (Novagen) was subjected to the same double enzyme digestion treatment;
[0082] The digested plasmid and gene were ligated overnight at 16°C;
[0083] Transform Escherichia coli DH5α the next day, and apply Kan (kanamycin, 50mg / L) resistant LB plates for screening;
[0084] The positive clones obtained by screening were extracted and sequenced after extraction of plasmids, and the plasmids with correct sequencing results were stored for later use.
Embodiment 3
[0085] Embodiment 3: transformation Escherichia coli BL21 (DE3)
[0086] The plasmid constructed in Example 2 was mixed with chemically competent cells BL21(DE3) (purchased from Invitrogen) and then kept in an ice bath for 20 minutes; heat-shocked at 42°C for 90 seconds; quickly added to SOC medium (purchased from Invitrogen), and incubated at 37°C for 1 Paint the panels after hours.
PUM
| Property | Measurement | Unit |
|---|---|---|
| chromatographic purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com