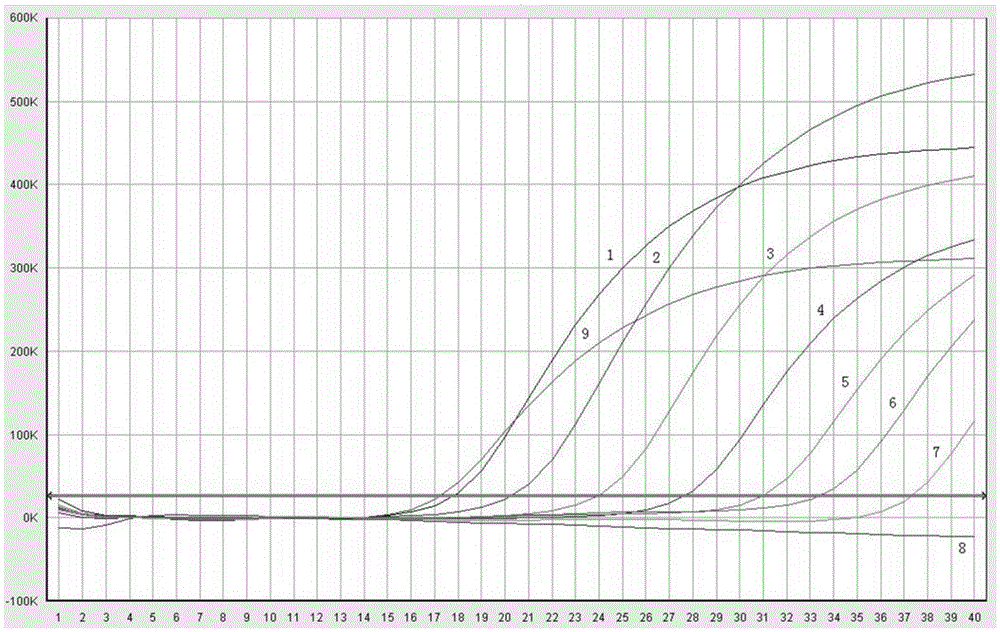
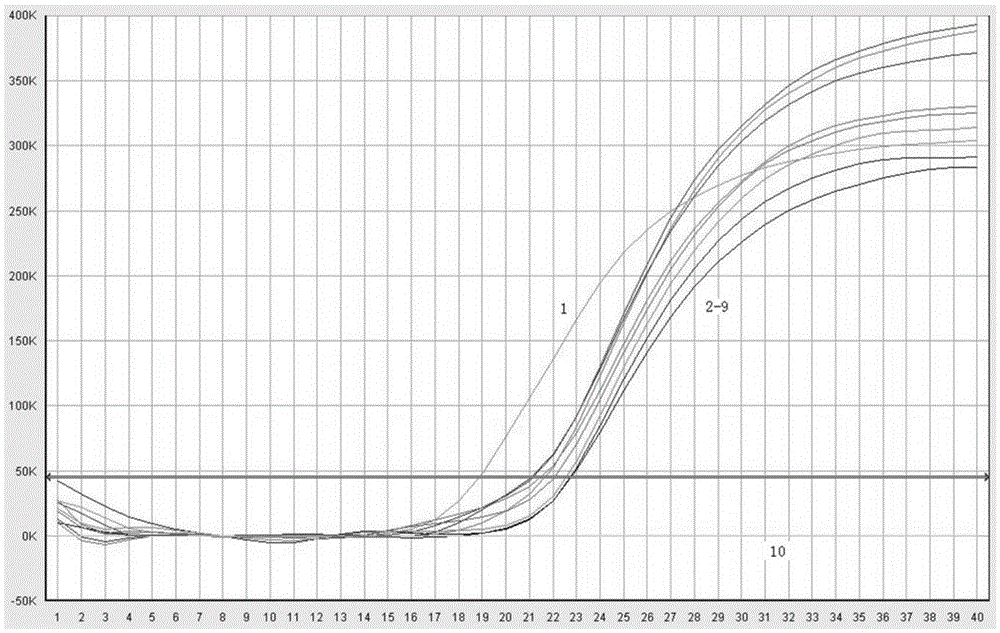
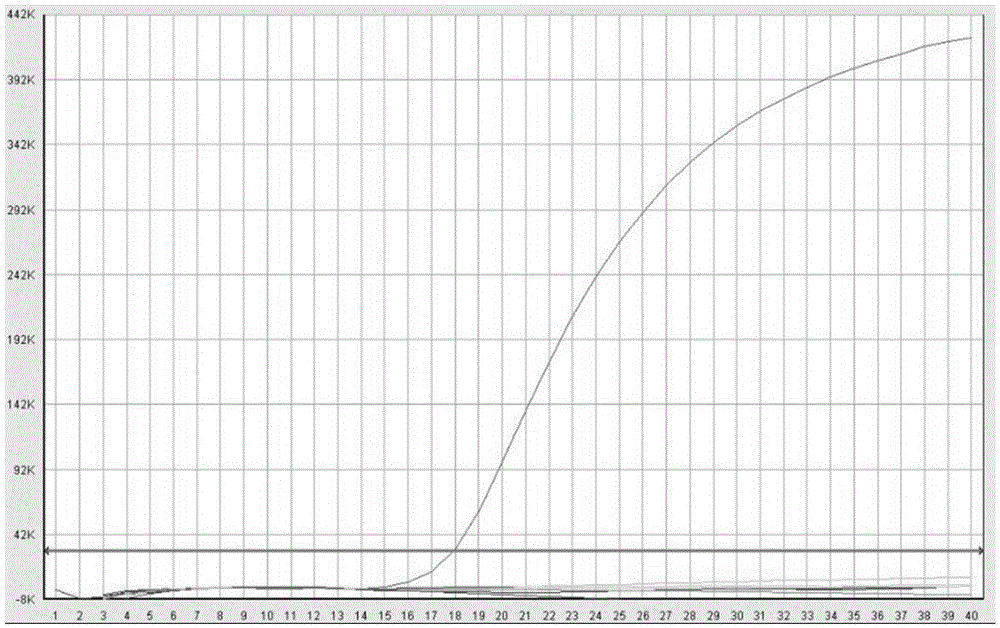
Real-time fluorescence PCR (polymerase chain reaction) primer and probe for identifying culex tritaeniorhynchus and application of real-time fluorescence PCR primer and probe
A real-time fluorescence and probe technology, applied in the field of insect identification, achieves high sensitivity, solves the problem of mosquito species identification, and is easy to operate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Primer Design:
[0041] According to the gene sequence of Culex tritaeniorhynchus COI (cytochromeoxidaseI) in GenBank (gene accession numbers AB690857, AB738091, HM638220, HQ398885, JQ728031, KC970299, KF407833, KM502254), the primers Culex-F and Culex-R, and the specific probe Tritaeniorhynchus were designed -P.
[0042] Fluorescence PCR upstream primer Culex-F: 5'-TTCAAGTAGTTTAGTAGAAAATGGAGC-3' (as shown in SEQ ID NO.1)
[0043] Fluorescent PCR downstream primer Culex-R: 5'-TGTAAAGAAAAAATAGCTAAATCAACTG-3' (as shown in SEQ ID NO.2)
[0044] Fluorescent PCR probe Tritaeniorhynchus-P: FAM-5'-ACAGTTTTATCCACCTCT-3'-MGB (as shown in SEQ ID NO.3)
Embodiment 2
[0045] Example 2 mosquito DNA extraction:
[0046] Mosquito DNA was extracted using commercial InsectDNAKit (OMEGA). The extracted DNA was tested directly on the machine or stored at -20°C.
Embodiment 3
[0047] The preparation of embodiment 3 standard plasmids
[0048] Referring to the literature DNAprimersforamplificationofmitochondrialcytochromecoxidasesubunitIfromdiversemetazoaninvertebrates (FolmerO, BlackM, HoehW, etal.MolMarBiolBiotechnol.1994,3(5):294-299) synthesis and amplification of COI gene upper and lower primers LCO1490 and HCO2198.
[0049] Culex tritaeniorhynchus DNA was extracted according to the instructions of InsectDNAKit (OMEGA).
[0050] The PCR reaction was performed using the commercialized ExTaq from Treasure Bio (Dalian) Co., Ltd. Add 5 μL of 10×Buffer, 4 μL of 2.5 mmol / LdNTP, 1 μL of 20 μmol / L upstream and downstream primers LCO1490 and HCO2198, 0.25 μL of ExTaq, 5 μL of DNA template, and supplemented with dHO in the PCR reaction tube. 2 0 to 50 μL. The reaction program was: pre-denaturation at 94°C for 3 minutes; 94°C for 30s, 45°C for 30s, 72°C for 1min, 5 cycles; 94°C for 30s, 51°C for 1mins, 72°C for 1min, 35 cycles; 72°C for 10min. PCR produc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com