A characteristic nucleotide sequence, nucleic acid molecular primer and method for quantitative detection of Cordyceps guangdong
A nucleotide sequence, Cordyceps in Guangdong technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as inability to quantitatively detect, achieve simple methods, short time, specificity Good results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Extraction of Genomic DNA of Cordyceps guangdong and Acquisition of α-tubulin Protein Coding Gene Sequence
[0019] Weigh 0.05g-0.15g Cordyceps guangdong sample and place it in a sterile mortar, pour liquid nitrogen into it and grind it quickly, select UNlQ-10 column type fungal genome DNA extraction kit [Sangon Bioengineering (Shanghai) Co., Ltd. Co., Ltd., No. B511375-0050], extracted Genomic DNA of Cordyceps guangdong, and purified Genomic DNA of Cordyceps guangdong with SanPrep Column DNA Gel Recovery Kit [Sangon Bioengineering (Shanghai) Co., Ltd., No. B518131-0050]. Genomic DNA of Cordyceps guangdong was obtained. Based on the α-tubulin protein-encoding gene family sequence in Genbank, specific nucleic acid molecular primers FL and RL (FL: 5`-TGGTAACGCCTGCTGGGA-3`, RL: 5`-YTCGCCGACGTACCAGTG-3` were provided by Shanghai Sangon Bioengineering Co., Ltd. Ltd Synthetics). Using the nucleic acid molecule primers FL and RL as primers, using the Genomic DNA of Cordyceps...
Embodiment 2
[0021] Fluorescent PCR Amplification with Specific Primers of Cordyceps guangdong
[0022] According to the α-tubulin protein coding gene (SEQ ID NO.1) of Cordyceps guangdong, a pair of sequences composed of 16-17 nucleotides, namely FS and RS, was designed by using the primer design software primer6, and the sequences are as follows:
[0023] FS: 5`-GCTCCGATTCGATGGC-3`; RS: 5`-TACAGCAGGCAGGTGGC-3`. Primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd. The sequence obtained by primer amplification is shown as SEQ ID NO.2, with a total of 234 bp bases.
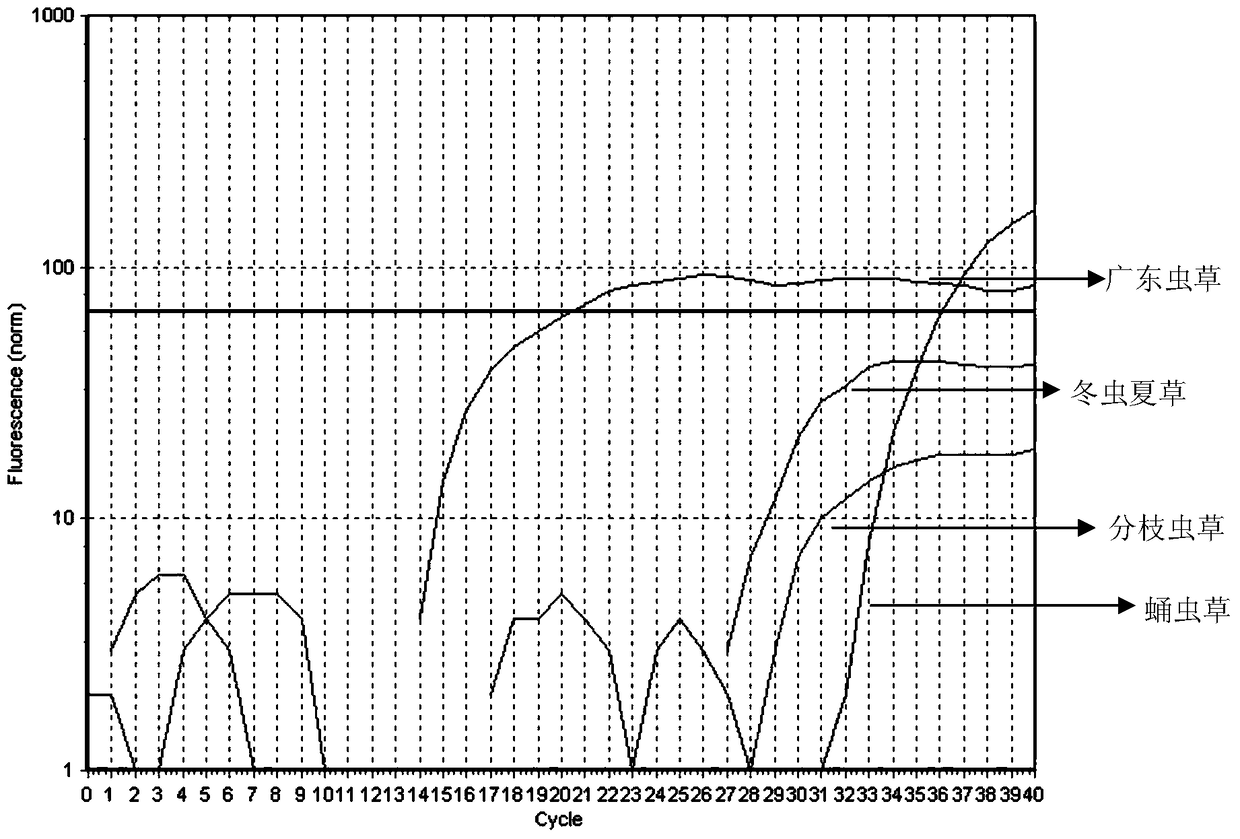
[0024] Using the SuperReal PreMix (SYBR Green) [Tiangen Biochemical Technology (Beijing) Co., Ltd., No. FP204] kit, carry out fluorescent quantitative PCR reaction, and use Cordyceps guangdong genomic DNA and Cordyceps sinensis, Cordyceps militaris, branch Cordyceps ramosa Genomic DNA as template. The total volume of the reaction system is 20 μL: 2×SuperReal PreMix 10.0 μl, FS (10 μmol / L) 0.5 μL, RS (10 μmol / L...
Embodiment 3
[0028] Detection of the initial concentration of DNA of Cordyceps guangdong
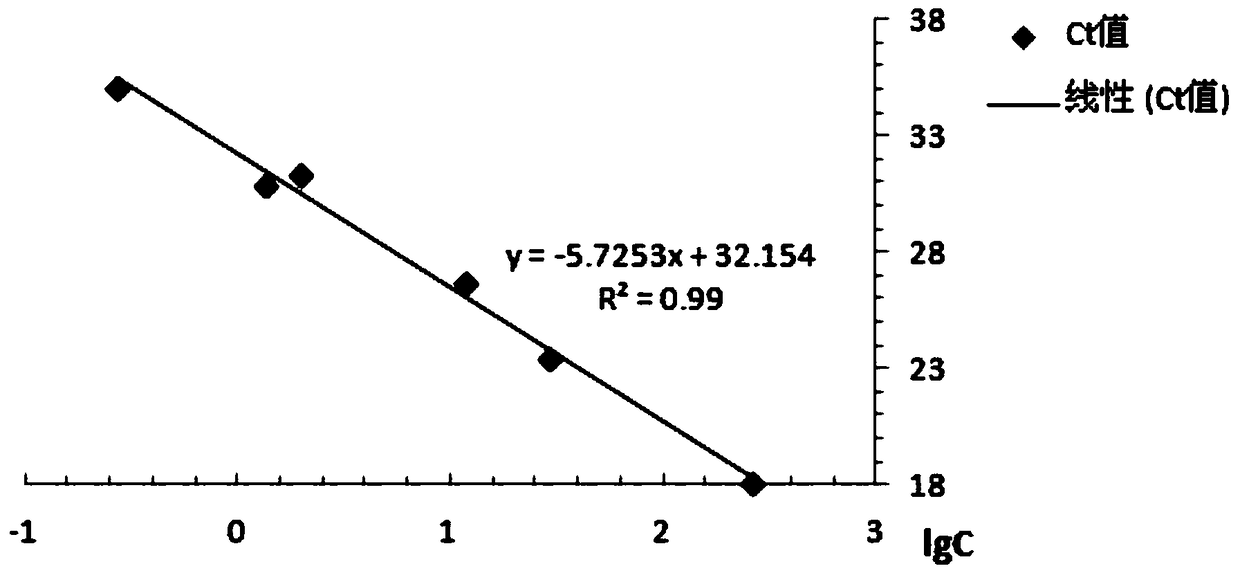
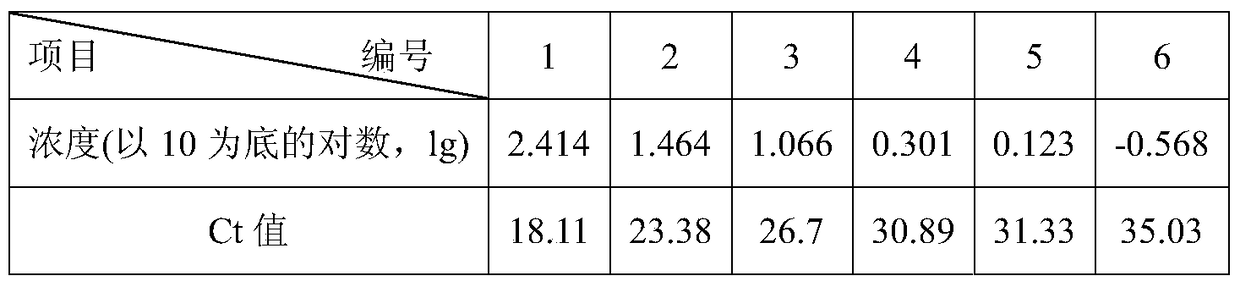
[0029] According to the known different concentrations of Cordyceps guangdong genomic DNA samples as templates, using FS and RS as primers to amplify according to the fluorescent quantitative PCR method of Example 2, the Ct value of the obtained Cordyceps guangdong α-tubulin protein coding gene fluorescent PCR (See Table 1), draw standard curve (see figure 2 ). The correlation coefficient is 0.990, the amplification efficiency is 99.5%, the linear range of Ct value is 18.11-35.03, and the regression equation is y=-5.7253x+32.154. According to the regression equation, the relative content of the tested samples of Cordyceps guangdong can be calculated.
[0030] Table 1 Ct values of different concentrations of Cordyceps guangdong DNA samples
[0031]
[0032]
[0033]
[0034]
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com